Understanding circadian rhythms in algae and fungi

Fungi, algae, and cyanobacteria might not complain about jet lag. But like humans, their physiologies adhere to a roughly 24-hour cycle of behavioral patterns in the absence of external cues. Organisms that experience recurring day and night cycles have evolved a biochemical oscillator or circadian clock. This clock determines which activities, from sleep to cellular metabolism, occur at biologically advantageous times.
Since nearly all organisms share a circadian rhythm, biologists have a plethora of genetically tractable opportunities to study it in the laboratory. Research has focused on how changes in transcribed genes can lead to expressions of different proteins throughout the 24-hour cycle. Cyclical or 'rhythmic' expression dictated by the circadian clock allows organisms to anticipate regular environmental changes by optimizing the complement of proteins present at any given time.
Unique capabilities offered by EMSL, the Environmental Molecular Sciences Laboratory, a DOE Office of Science user facility at Pacific Northwest National Laboratory (PNNL) in Richland, Wash., provide biologists with the opportunity to study with unprecedented detail how proteins and metabolic pathways change over the course of a day. The DOE's Office of Biological and Environment Research (BER)'s interest in improving the robustness and resilience of biofuel plants and fungi has motivated several recent investigations into drivers of circadian rhythm.
From cyanobacteria to circadian rhythms
EMSL's involvement in circadian rhythm research has its origins in EMSL's Grand Challenges Program. In 2005, Himadri Pakrasi, professor of biology at Washington University in St Louis, worked with David Koppenaal, PNNL chief technology officer, and together they used resources made available by that program to start developing systems-level models of cyanobacteria. Pakrasi responded to an EMSL call for proposals in 2010, which led to the Synthetic Biology Research Campaign and a series of publications about how to optimize the photosynthetic microorganisms genetically for biomass production.
"Cyanobacteria are the only prokaryotes known for sure that have circadian rhythm just like us," says Pakrasi.
The oxygen-generating photosynthetic microbes are major contributors to the global carbon and nitrogen cycles, and implementation of daily rhythms by a circadian clock to control cellular physiology allows these organisms to adapt to environmental changes including light and temperature. Pakrasi points out that while researchers studying cyanobacteria have long observed rhythmic daily processes, the research community thought the organisms "lacked the complexity necessary for biological clocks." However, observations of oscillations in cellular functions such as nitrogen fixation have proven that cyanobacteria fulfil the criteria for a circadian oscillator.
Pakrasi and colleagues continue their work to understand how daily processes in cyanobacteria are controlled, including the mechanisms that regulate fast cell growth, light harvesting, and cellular redox control. Their work makes use of comparative proteomics, comparing changes in the entire complement of proteins expressed by an organism. Such measurements require the specialized instrumentation and skill sets of EMSL.
"Proteomics and metabolomics measurements are two sets of data a lot of institutions just can't do," says William Cannon, PNNL systems biologist.
Not just genes
Biologists Jennifer Hurley, Rensselaer Polytechnic Institute, and Jay Dunlap, Geisel School of Medicine Dartmouth, learned about EMSL's proteomics capabilities from the user facility's lead biologist Scott Baker. They submitted a proposal to investigate the proteome of a fungus with a circadian clock like that in humans.
In all living cells, DNA is transcribed into RNA in the nucleus. The transcribed RNA—the transcriptome—travels out of the nucleus into the cytoplasm, where it is translated into proteins. The simplest explanation for circadian rhythms is that if more RNA is transcribed at one time of day than another, then there should be more of certain proteins at certain times of day. But that is not always true.
"We'd started to learn that we can't trust what we saw in the transcriptome to reliably predict the proteome," says Hurley.
The researchers sent samples of Neurospora crassa, an organism valued for its ability to produce cellulose for biofuels, to EMSL. There, Erika Zink and Sam Purvine broke the cells open, extracted the proteins, and analyzed the samples using advanced mass spectrometry techniques.
"We're finding out the relative quantities of pairs of proteins, and looking for general trends that change over time," says Purvine.
For circadian rhythms, that means tracking which protein levels change significantly in cells pulled from growth chambers every two hours over a 48-hour period.
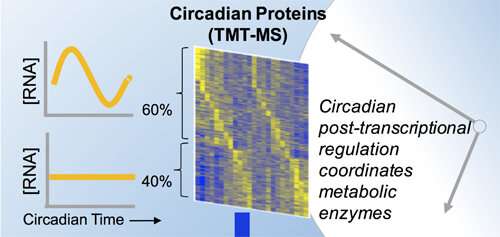
Hurley and Dunlap found a surprising pattern in the proteomic data. Of the proteins that rose and fell over a 24-hour period in Neurospora's life, only 60 percent corresponded to changes in the transcriptome. The rest of the proteins varied despite corresponding RNA that did not oscillate.
"That's 40 percent of the proteome you'd never know was rhythmic if you only looked at the transcriptome," says Dunlap.
The researchers' first hypothesis for the lack of correspondence between proteome and transcriptome was that protein degradation follows a circadian rhythm. Instead, they found the answer in the rate of translation.
"At some time of day, cells make proteins faster from RNA than at other times," says Dunlap.
The timing of these proteins leads to coordination of different metabolic pathways that are vital to the cell's overall function.
The work highlighted the necessity of studying the proteome to understand how the circadian clock regulates an organism's metabolic output. Future research will investigate how metabolic feedback can alter circadian rhythm regulation; and on the long timescale, whether the circadian clock is an evolutionary adaptation to metabolism.
Unexpected pathways
Like Neurospora, the single-celled green algae Chlamydomonas reinhardtiis holds potential for creating biofuels. Biochemist Sabeeha Merchant, University of California at Los Angeles (UCLA), studies how the molecular biology of the organism responds to light and dark cycles; specifically, how a cell decides when to divide in two. She had worked on several other publications with EMSL staff and sought the user facility's expertise to learn about Chlamydomonas' proteome.
Merchant and UCLA project scientist Daniela Strenkert cultivated Chlamydomonas on a 12-hour light-dark cycle to establish a synchronous culture of cells. The cycle ensured each of millions of cells divided once every 24 hours. The experimental system allowed the researchers to observe the sequential order of events and pathways during the process of cell division, amplified millions of times.
Merchant, Strenkert, and their team extracted RNA from the cells every two hours over the course of a diurnal cycle—a 24-hour light/dark cycle caused by the daily rotation of the Earth—and determined the transcriptome.
"You don't know if there's any protein present when you see the RNA," says Merchant. "You need to validate it by looking at the proteome."
With proteomics data provided by Purvine and Mary Lipton at EMSL, Merchant analyzed how the transcriptome compared to measurements of pigments, metabolites, and physiological changes. The researchers found that 85 percent of the Chlamydomonas genome was expressed differently over the course of the day to coordinate cellular growth before cell division. The timing of changes in the genes was dictated by their biological function; for example, a sigma factor that promotes transcription in the chloroplast increased in the dark, right before the light went on.
Metabolic pathways, too, followed a diurnal cycle. As expected for any photosynthetic cell, Chlamydomonas stored carbon in the form of glucose or starch during the day and broke the starch down at night to yield pyruvate. The researchers expected the cells would then extract more energy from pyruvate by its complete oxidation coupled to respiration. In respiration, cofactor molecules (NADH) produced during the oxidation of pyruvate are regenerated by oxygen allowing an organism to extract even more energy.
But to the researchers' surprise, Chlamydomonas did not engage in respiration in the dark. Rather, cells have another option for metabolizing pyruvate in the absence of oxygen. In fermentation, an organism metabolizes pyruvate via conversion to lactate.
"This is the same process that occurs in our muscles when we run out of oxygen," says Merchant. "We thought that giving them enough oxygen meant they would always use respiration."
Even with abundant oxygen, the algae engaged a fermentation pathway.
The team proposed possible explanations for the surprising pathway: the cells might not require much energy in the dark and speed may take precedence over energy generation. Pyruvate metabolism by fermentation may be faster than pyruvate metabolism coupled to respiration. Or, the algae may deliberately choose fermentation so that they might provide fermentation products for other microbes in their environment as a form of cooperative interaction.
The detailed proteomic and transcriptomic data sets originating from these projects will be a valuable resource as the research community explores how circadian clocks lead to coordination of different metabolic pathways that are vital for a cell's overall function.
More information: Jennifer M. Hurley et al. Circadian Proteomic Analysis Uncovers Mechanisms of Post-Transcriptional Regulation in Metabolic Pathways, Cell Systems (2018). DOI: 10.1016/j.cels.2018.10.014
Daniela Strenkert et al. Multiomics resolution of molecular events during a day in the life of Chlamydomonas, Proceedings of the National Academy of Sciences (2019). DOI: 10.1073/pnas.1815238116
Michelle Liberton et al. Phycobilisome truncation causes widespread proteome changes in Synechocystis sp. PCC 6803, PLOS ONE (2017). DOI: 10.1371/journal.pone.0173251
Jingjie Yu et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2, Scientific Reports (2015). DOI: 10.1038/srep08132
Jia Guo et al. Proteome-wide Light/Dark Modulation of Thiol Oxidation in Cyanobacteria Revealed by Quantitative Site-specific Redox Proteomics, Molecular & Cellular Proteomics (2014). DOI: 10.1074/mcp.M114.041160
Journal information: Cell Systems , Proceedings of the National Academy of Sciences , PLoS ONE , Scientific Reports , Molecular & Cellular Proteomics
Provided by Environmental Molecular Sciences Laboratory