This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Hidden mechanisms behind hermaphroditic plant self-incompatibility revealed
A new study presents an evolutionary-biophysical model that sheds new light on the evolution of the collaborative non-self recognition self-incompatibility, a genetic mechanism in plants that prevents self-fertilization and promotes cross-fertilization. The innovative model introduces promiscuous molecular interactions as a key ingredient, enhancing our understanding of genetic diversity and evolution in hermaphroditic plants.
A study led by Dr. Tamar Friedlander and her team at The Robert H. Smith Institute of Plant Sciences and Genetics in Agriculture at the Hebrew University, in collaboration with Prof. Ohad Feldheim from the Einstein Institute of Mathematics at the Hebrew University has developed an evolutionary-biophysical model that sheds new light on the evolution of collaborative non-self recognition self-incompatibility in plants.
The study, appearing in Nature Communications, introduces a novel theoretical framework that incorporates promiscuous molecular interactions, which have been largely overlooked by traditional models.
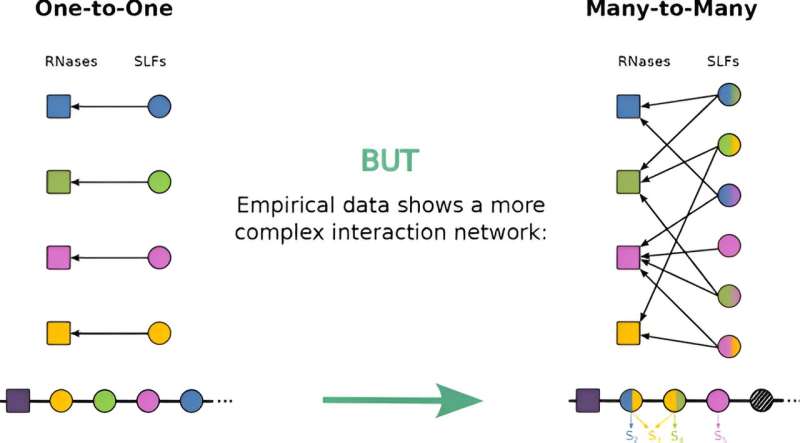
Self-incompatibility (SI) is a widespread biological mechanism in plants having both male and female reproductive organs, that prevents self-fertilization and promotes genetic diversity. Under this mechanism, fertilization relies on the specific recognition between highly diverse proteins: the RNase (female determinant) and the SLF (male determinant).
The interaction between these proteins ensures that plants are only compatible with non-self mates, thus maintaining a diverse gene pool.
The new model proposed by Dr. Friedlander and her team represents a significant advancement in understanding the evolutionary dynamics of self-incompatibility proteins. By allowing for promiscuous interactions—where interactions with unfamiliar partners are likely—and for multiple distinct partners per protein, the model aligns more closely with empirical findings than previous models that assumed only one-to-one interactions.
This promiscuity enables a flexible interaction pattern between male and female proteins, offering new insights into how these proteins evolve and interact over generations.
"Our research shows that the ability of proteins to engage in promiscuous interactions is crucial for the long-term evolutionary maintenance of self-incompatibility systems," explained Dr. Friedlander.
"We propose that the default state of this system is that recognition is likely and evolutionary pressure is needed to avoid it, in contrast to what was previously thought. This flexibility not only helps in maintaining genetic diversity but also suggests that similar mechanisms could be operating in other biological systems."
The study also reveals how populations of these plants spontaneously organize into distinct compatibility classes, ensuring full compatibility across different classes while maintaining incompatibility within the same class.
The model predicts various evolutionary paths that could lead to the formation or elimination of these compatibility classes based solely on point mutations. The dynamic balance between the emergence and decay of these classes, which provides a sustainable model of evolution, was analyzed by the researchers using a mixture of empirical and theoretical tools borrowed from the field of statistical mechanics in physics.
"These insights from our study have profound implications not only for plant biology but also for understanding the fundamental principles of molecular recognition and its impact on the evolution of biological networks," Dr. Friedlander added. "Our findings could also help in the conservation of plant biodiversity."
This research, which highlights the role of promiscuous and multi-partner molecular interactions, is likely to inspire seeking these two elements in additional biological systems, and help in explaining the evolution of various complex molecular networks. It enriches the understanding of plant biology and has broader implications for deciphering the evolution of biological networks and managing biodiversity.
More information: Keren Erez et al, The role of promiscuous molecular recognition in the evolution of RNase-based self-incompatibility in plants, Nature Communications (2024). DOI: 10.1038/s41467-024-49163-7
Journal information: Nature Communications
Provided by Hebrew University of Jerusalem