Small RNA renders bacteria more resistant to antibiotics

Many soil bacteria are resistant to antibiotics by nature. A new mechanism for regulating that resistance has been identified by researchers at Ruhr-Universität Bochum. In the journal mBio, published online on 13 November 2018, the team headed by Jessica Borgmann from the Chair of Microbial Biology describes a small RNA molecule that significantly affects antibiotic resistance as well as other processes inside the cell.
"Soil bacteria that are resistant to antibiotics have a competitive advantage over rival microorganisms that excrete such antibiotics," explains Professor Franz Narberhaus, Head of the Chair of Microbial Biology. Agrobacterium tumefaciens, a soil bacterium that causes plant tumours, is known for its tolerance of beta lactam antibiotics, i.e. penicillins. The researchers have now explained the mechanism underlying this resistance. They were primarily interested in a small RNA molecule that regulates various processes in Agrobacterium.
Less tolerant without small RNA
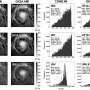
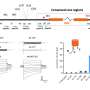
Together with Professor Julia Bandow's Applied Microbiology research group, Borgmann's team analysed how protein patterns inside the cell change when the small RNA is missing. The result: the bacteria produced considerably smaller volumes of the enzyme that degrades penicillins. The researchers, moreover, tested what happened when they added ampicillin to the bacterial culture. The strain without small RNA was one hundred times more sensitive to the antibiotic than the natural strain.
"Accordingly, small RNA appears to be the reason why the natural strain is so tolerant to some antibiotics," says Jessica Borgmann. "We assume that the main function of this molecule is to ensure an advantage for bacteria that compete with rival microorganisms."
Interference with different metabolic pathways
Conducting further analyses, the Bochum-based researchers found out that small RNA manipulates numerous processes, those including biosynthesis of the bacterial cell wall. If the bacteria lacked this molecule, however, they formed more plant tumours than the natural strain. In nature, as the researchers suspect, the two uses of cellular resources are balanced against each other: higher antibiotic resistance versus higher virulence in the forming of tumours.
"The results suggest a complicated interaction of different metabolic pathways," concludes Franz Narberhaus. "Apparently, what counts in nature is to coordinate several pathways, in order to ensure fitness under changing conditions."
Small RNAs as efficient regulators
Short RNA molecules are very efficient in switching cellular processes on and off, according to the requirements. Their synthesis is considerably faster and more energy-saving for the cells than the production of protein regulators.
In follow-up studies, the researchers from Bochum intend to figure out the precise mechanism deployed by RNA molecules to manipulate different processes. Moreover, they also wish to examine whether antibiotic resistance in other soil bacteria is based on a similar principle and if such resistance can be transferred to other types of bacteria.
More information: Jessica Borgmann et al. A Small Regulatory RNA Controls Cell Wall Biosynthesis and Antibiotic Resistance, mBio (2018). DOI: 10.1128/mBio.02100-18
Journal information: mBio
Provided by Ruhr-Universitaet-Bochum