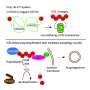
Team successfully analyzes the structure of the protein FAT10 toward potential cancer therapy
FAT10 is a small protein with a huge effect. Its attachment to a target protein is a signal for its degradation. FAT10 is a marking system for degradation that seems to be inefficient. In contrast to its biological competitor, ubiquitin, which is recycled, FAT10 is degraded along with its target protein, which appears wasteful at first glance.
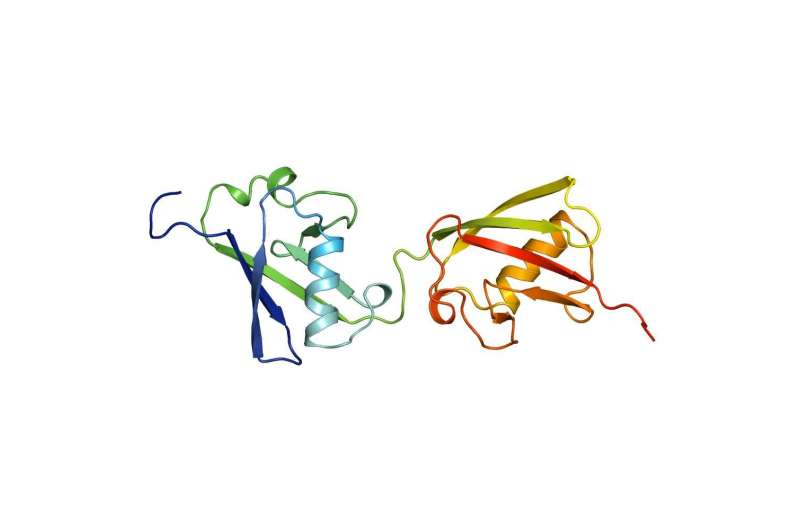
So why does this seemingly inefficient FAT10-system exist at all? University of Konstanz immunologist Marcus Groettrup and his team have been carrying out research on FAT10 for many years. Now, they report the high-resolution structure of FAT10. This success was enabled through another achievement. In collaboration with Dr. Annette Aichem from the Biotechnology Institute Thurgau (BITg), Konstanz chemist Professor Christine Peter and structural biologist Dr. Silke Wiesner from the University of Regensburg, the team developed a molecular technique to produce stable and highly-concentrated FAT10 with a high degree of purity. As a consequence, the researchers could carry out a structure analysis of FAT10 via X-ray crystallography and magnetic resonance spectroscopy. The results have been published in the current issue of Nature Communications.
"We found a mechanism in FAT10 that is entirely different from ubiquitin. This mechanism is very interesting for the entire ubiquitin field," says Groettrup. In contrast to ubiquitin with one domain, FAT10 has two domains, i.e. folds that enable the proteins to function. The team has now discovered that both domains are connected by a flexible linker and their folding is significantly less compact than in ubiquitin. Importantly, FAT10 has a short, disordered extension.
Experiments show that the loose folding and the disordered extension of the FAT10 protein have an important regulatory function that actually simplifies the degradation of the target molecule. As a result, no complicated upstream processes of partial substrate unfolding are necessary, as in the ubiquitin-system, to have the enzyme 26S proteasome, which actually carries out the degradation, spring into action. This is superfluous due to the loose folding of the FAT10 domains. The enzyme targets the disordered part and can easily unfold the two flexible and loosely folded domains, and consequently can pull apart the attached target protein and degrade it.
It had been already known before that ubiquitin and FAT10 have entirely different binding properties. That is all the more surprising, given the fact that the folding of both proteins' domains is based on the same structure. The findings of the research team now explain why: The surfaces of FAT10 and ubiquitin are entirely different, and they determine which proteins are bound. Consequently, both marking systems have different interaction partners. FAT10 is predominantly found in inflammatory tissue. "Without inflammation, it virtually is not present at all," says Groettrup. In his view it is an advantage that the marking system is destroyed along with the protein. His hypothesis: Nature counts on irreversibility in this context so the protein degradation cannot be reversed.
Noting another interesting fact about FAT10, Groettrup says, "In 13 different types of cancer, it is found to an extended degree. We can conclude that FAT10 is advantageous for the cancer cells' survival and growth." If FAT10, for example, links to proteins that suppress tumour growth, they are degraded, and the tumour cell can grow more easily. For such cases, it is conceivable to develop a cancer drug inhibiting the enzymes that link FAT10 and the target proteins. "Talking about practical implementation, our findings actually provide grounds for hope," concludes Groettrup.
More information: Annette Aichem et al, The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation, Nature Communications (2018). DOI: 10.1038/s41467-018-05776-3
Journal information: Nature Communications
Provided by University of Konstanz