Snapshot of DNA repair
DNA is like the computer code of the body, and it must be preserved for our bodies to survive. Yet, as cells grow and change, DNA is vulnerable to defects, especially double strand breaks (DSBs). In fact, DSBs regularly occur throughout one's lifetime. The DNA is protected, however, by DNA repair machinery. A new study from scientists at Osaka University and The University of Tokyo describes the crystal structure of a crucial part of this machinery, the binding of RNF168 to ubiquitylated proteins and find a unique binding not seen in other ubiquitylated protein interactions.
To activate the DNA repair machinery, DSBs stimulate ubiquitylation. RNF168 binds to polyubiquitylated proteins and accumulates at the damaged DNA as part of the repair process.
Ubiquitylation describes the binding of the protein ubiquitin to another protein. Ubiquitin will then further bind to itself through one of its seven lysine amino acid residues to form a ubiquitin chain. RNF168 binds with high specificity to ubiquitin chains linked at lysine 63.
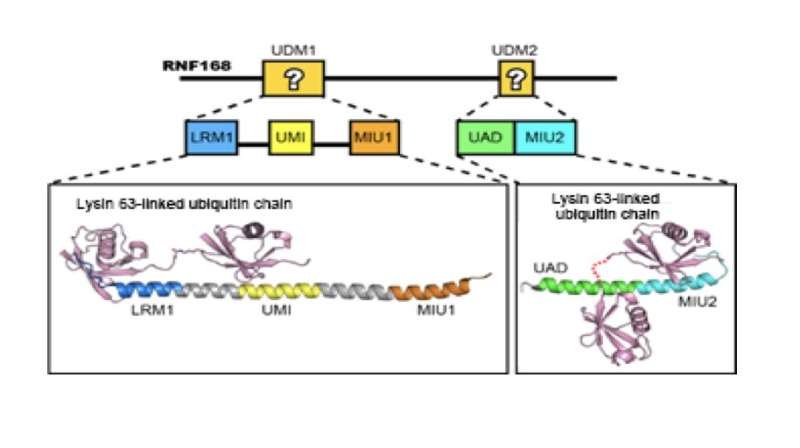
Because the binding of RNF168 to lysine 63 chains is imperative for the recruitment of other DNA repair proteins, the scientists investigated the molecular interactions that assure this binding. In particular, they were interested in two RNF168 domains appropriately named ubiquitin-dependent DSB recruitment module (UDM)1 and UDM2.
Crystal structures were made of each RNA168 domain bound to a lysine 63 chain two ubiquitin molecules long. The structures revealed that the RNF168-lysine 63 chain interaction is stabilized by many unique hydrogen bonds and hydrophobic interactions not seen in other ubiquitin lysine chains. Furthermore, UDM1 and UDM2 each folded as single α–helices to bind to the distal and proximal ubiquitins, but the specificity for lysine 63 chains by RNF168 depended on the interaction with the distal ubiquitin. This, the study noted, is in striking contrast to other interactions where proximal ubiquitin defines the linkage specificity.
Using the crystal structures, the researchers could identify which regions in each UDM bound to the two ubiquitin molecules: LRM1 and UMI in UDM1, and UAD and MIU2 in UDM2. However, the conditions for binding were different. For UDM1, the specificity to lysine 63 chains depended on the distance and orientation between LRM1 and UMI while RNF168 was bound to ubiquitin. On the other hand, for UDM2, it was the space between the two ubiquitin molecules while RNF168 was bound to ubiquitin. Subsequent mutant experiments revealed that UDM2 binding was essential for RNF168 recruitment, while UDM1 was auxiliary.
More information: Tomio S. Takahashi et al. Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168, Nature Communications (2018). DOI: 10.1038/s41467-017-02345-y
Journal information: Nature Communications
Provided by Osaka University