Choosing the right substrate for the right function
Scientists at Tokyo Institute of Technology have discovered a unique molecular mechanism responsible for the substrate preference of ubiquitin-specific proteases.
Ubiquitin is a small, highly conserved, eukaryotic regulatory protein that can be attached to Lys residues on intracellular proteins via isopeptide bonds. Ubiquitin can also be attached to one of seven Lys residues of other ubiquitin moieties forming poly-ubiquitin chains on target proteins. Depending on which Lys in ubiquitin is used to form poly-ubiquitin chains, distinct signaling functions take place. Thus, Lys48-linked polyubiquitination, which is the most abundant in the cell, results in proteasomal degradation of target proteins, whereas Lys63-linked polyubiquitination regulates multiple cell activities such as protein trafficking and DNA repair. Ubiquitination is reversible as the formed isopeptide bonds can be cleaved by deubiquitinating enzymes which sometimes exhibit different preferences for Lys48- and Lys63-linked ubiquitin chains. However, the molecular mechanisms underlying the substrate specificity of deubiquitinases toward Lys48-linked ubiquitin chains remain largely unknown.
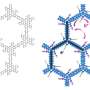
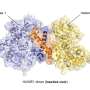
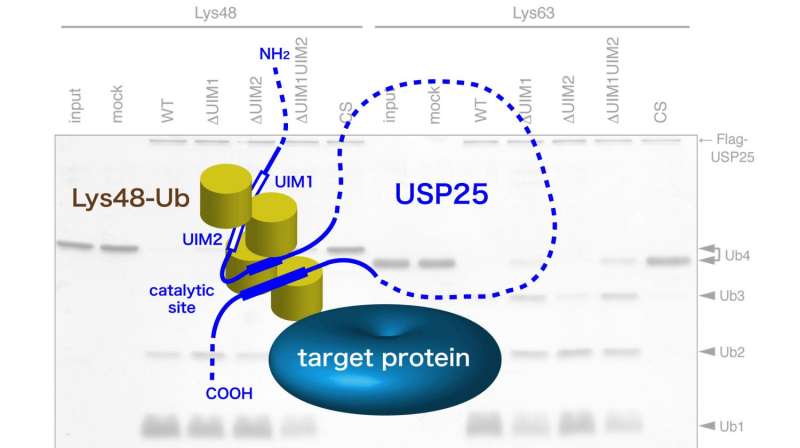
Researchers at Tokyo Institute of Technology led by Professor Masayuki Komada addressed this problem by investigating the function of ubiquitin-specific protease (USP)25, which in this study was found to favor the Lys48 linkage for cleavage. USP25 contains two tandem ubiquitin-interacting motifs (UIMs) representing 20-amino-acid α-helices (Figure). To test whether UIM-mediated binding results in substrate preference, these scientists generated a panel of USP25 mutants carrying substitutions of functional amino acids in UIMs and analyzed their interaction with and catalytic activity toward Lys48- or Lys63-linked ubiquitin chains. It appeared that the tandem UIMs not only enhanced the isopeptidase activity of USP25 but also determined its preferential cleavage of Lys48-linked ubiquitin chains. Preferential binding of the UIMs to Lys48-linked ubiquitin chains seems to play a role, as swapping of the UIMs to those of other proteins with binding specificity for Lys48-linked chains significantly shifted USP25 preference from the Lys48 towards Lys63 linkage, suggesting the importance of UIM's ubiquitin chain-binding specificity for USP25's substrate specificity.
These findings present the first example of deubiquitinases in which the substrate preference toward Lys48-linked ubiquitin chains is determined by UIMs. The scientists suggest that UIMs play a role in the catalytic mechanism by keeping Lys48-linked ubiquitin chains in close proximity to the active site of the enzyme (Figure). Future structural studies of USP25 complexed with a Lys48-linked ubiquitin chain should determine whether this is indeed the case.
More information: Kohei Kawaguchi et al, Tandem UIMs confer Lys48 ubiquitin chain substrate preference to deubiquitinase USP25, Scientific Reports (2017). DOI: 10.1038/srep45037
Journal information: Scientific Reports
Provided by Tokyo Institute of Technology