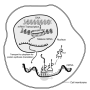
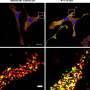
Methyl marks on RNA discovered to be key to brain cell connections
Methyl chemical groups dot lengths of DNA, helping to control when certain genes are accessible by a cell. In new research, UCLA scientists have shown that at the connections between brain cells—which often are located far from the central control centers of the cells—methyl groups also dot chains of RNA. This methyl markup of RNA molecules is likely key to brain cells' ability to quickly send signals to other cells and react to changing stimuli in a fraction of a second.
To dictate the biology of any cell, DNA in the cell's nucleus must be translated into corresponding strands of RNA. Next, the messenger RNA, or mRNA—an intermediate genetic molecule between DNA and proteins—is transcribed into proteins. If a cell suddenly needs more of a protein—to adapt to an incoming signal, for instance—it must translate more DNA into mRNA. Then it must make more proteins and shuttle them through the cell to where they are needed. This process means that getting new proteins to a distant part of a cell, like the synapses of neurons where signals are passed, can take time.
Research has recently suggested that methyl chemical groups, which can control when DNA is transcribed into mRNA, are also found on strands of mRNA. The methylation of mRNA, researchers hypothesize, adds a level of control to when the mRNA can be translated into proteins, and their occurrence has been documented in a handful of organs throughout the bodies of mammals. The pattern of methyls on mRNA in any given cell is dubbed the "epitranscriptome."
UCLA and Kyoto University researchers mapped out the location of methyls on mRNA found at the synapses, or junctions, of mouse brain cells. They isolated brain cells from adult mice and compared the epitranscriptome found at the synapses to the epitranscriptomes of mRNA elsewhere in the cells. At more than 4,000 spots on the genome, the mRNA at the synapse was methylated more often. More than half of these spots, the researchers went on to show, are in genes that encode proteins found mostly at the synapse. The researchers found that when they disrupted the methylation of mRNA at the synapse, the brain cells didn't function normally.
The methylation of mRNA at the synapse is likely one of many ways that neurons speed up their ability to send messages, by allowing the mRNA to be poised and ready to translate into proteins when needed.
The levels of key proteins at synapses have been linked to a number of psychiatric disorders, including autism. Understanding how the epitranscriptome is regulated, and what role it plays in brain biology, may eventually provide researchers with a new way to control the proteins found at synapses and, in turn, treat disorders characterized by synaptic dysfunction.
More information: Daria Merkurjev et al. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts, Nature Neuroscience (2018). DOI: 10.1038/s41593-018-0173-6
Journal information: Nature Neuroscience
Provided by University of California, Los Angeles