New beacons light up the interior
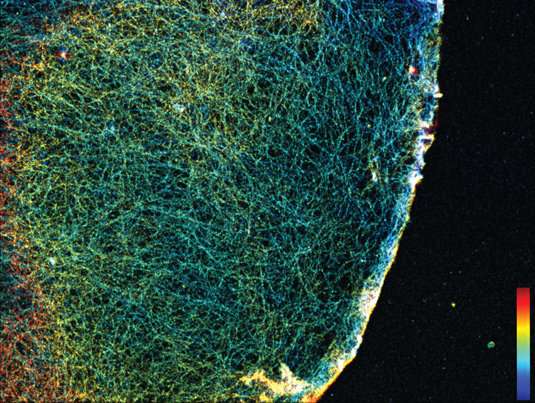
LMU's Ralf Jungmann develops modes of microscopy that can resolve cellular structures with dimensions on the order of nanometers. He has now succeeded in imaging actin networks in cells in greater detail than before.
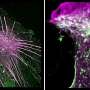
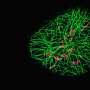
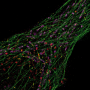
Ralf Jungmann, Professor of Experimental Physics at LMU and head of the research group Molecular Imaging and Bionanotechnology at the Max Planck Institute of Biochemistry (Martinsried), is engaged in the development of innovative modes of microscopy that make it possible to visualize intracellular processes at the single-molecule level. His approach is based on the use of fluorescent markers tagged with short single-stranded DNAs that define their binding specificity. These markers recognize their targets by binding to complementary DNA sequences attached to antibodies that interact specifically with individual cellular proteins. This strategy, aptly known as DNA-PAINT, allows one to specifically "address" a plethora of cellular proteins at once. Now, a team led by Jungmann, with Thomas Schlichthärle as first author, reports the first use of small proteins called 'affimers," in place of the bulkier antibodies, to visualize the actin networks in cells at even higher resolution. The work was carried out in collaboration with the groups led by Darren Tomlinson and Michelle Peckham at the University of Leeds as well as Jonas Ries from the European Molecular Biology Lab (EMBL) in Heidelberg. The new study appears in the journal Angewandte Chemie.
The goal of super-resolution microscopy is to visualize cellular structures and processes at the single-molecule level, with molecules being only a few nanometers in size. The antibodies so far used to detect cellular proteins are quite large – three to four times larger than the proteins to which they bind. This also holds true for the DNA-tagged antibodies employed to visualize target proteins in experiments with DNA-PAINT. The difference between the resolution provided by DNA-PAINT and the dimensions of the antibody-target complex essentially means that the fluorescent signal indicates the position of the antibody and not that of the protein to which it is bound. This problem can be solved and more precise and informative data can be obtained by developing smaller markers that offer the same level of recognition specificity.
In their latest project, Jungmann and his colleagues have turned to affimers for this purpose. Affimers are protein binders that are ten times smaller than traditional antibodies, but are still capable of specifically recognizing and binding to defined protein species. They are produced and displayed on the surfaces of bacterial viruses, and can be readily identified and purified. By modifying these affimers by site-specific attachment of a DNA strand of defined sequence, the researchers were able to clearly visualize single actin filaments in cells in three dimensions by means of DNA-PAINT. Up to now, this has required the use of extremely elaborate microscopy techniques and highly specialized markers. By combining DNA-PAINT with these novel affimers, it is now possible to achieve this level of resolution with a standard microscopy set-up and reagents that are easy to produce, says Thomas Schlichthärle. And Ralf Jungmann adds: Since affimer reagents directed against different proteins are relatively easy to make in the test tube, it should be possible in the future to use them to label large sets of proteins in cells. In combination with DNA-PAINT, this would allow us to visualize complete signaling pathways involving hundreds of different protein species."
More information: Thomas Schlichthaerle et al. Site-specific labeling of Affimers for DNA-PAINT microscopy, Angewandte Chemie International Edition (2018). DOI: 10.1002/anie.201804020
Journal information: Angewandte Chemie International Edition , Angewandte Chemie
Provided by Ludwig Maximilian University of Munich