Visualizing single molecules in whole cells with a new spin
Cell biologists traditionally use fluorescent dyes to label and visualize cells and the molecules within them under a microscope. With different super-resolution microscopy methods, they can even light up single molecules and their complex interactions with one another. However, the microscopy hardware that allows them to do this is highly specialized and expensive and hence, relatively rare in laboratories around the world, and the operation of such microscopes is daunting, as it requires unique skills.
Ralf Jungmann, Ph.D., an alumnus of Harvard's Wyss Institute for Biologically Inspired Engineering and currently a Professor at the Ludwig Maximilian University (LMU) and the Max Planck Institute (MPI) of Biochemistry in Germany and Wyss Institute Core Faculty member Peng Yin, Ph.D., have been developing DNA-PAINT, a powerful molecular imaging technology that involves transient DNA-DNA interactions to accurately localize fluorescent dyes with super-resolution. However, although the researchers have demonstrated DNA-PAINT's potential by visualizing single biomolecules, such as proteins, in fixed cells at a fixed close distance, the technology could not yet be applied to investigate molecules deep inside of cells.
Now, Jungmann's and Yin's teams jointly report a solution to overcome this limitation. In their new study, they adapted DNA-PAINT technology to microscopes that are widespread among cell biology laboratories, called confocal microscopes, and that are used by researchers to image whole cells and thicker tissues at lower resolution. The MPI/Wyss Institute team demonstrates that the method can visualize a variety of different molecules, including combinations of different proteins, RNAs and DNA throughout the entire depth of whole cells at super-resolution. Published in Nature Communications, the approach could open the door for detailed single-molecule localization studies in many areas of cell research.
The DNA-PAINT approach attaches a DNA "anchor strand to the molecule of interest. Then a dye-labeled DNA "imager strand" with a complementary sequence transiently attaches to the anchor and produces a fluorescent signal, which occurs as a defined blinking event at single molecular sites. Because "blinking" is precisely tunable, molecules that are only nanometers apart from each other can be distinguished—at the higher resolution end of super-resolution.
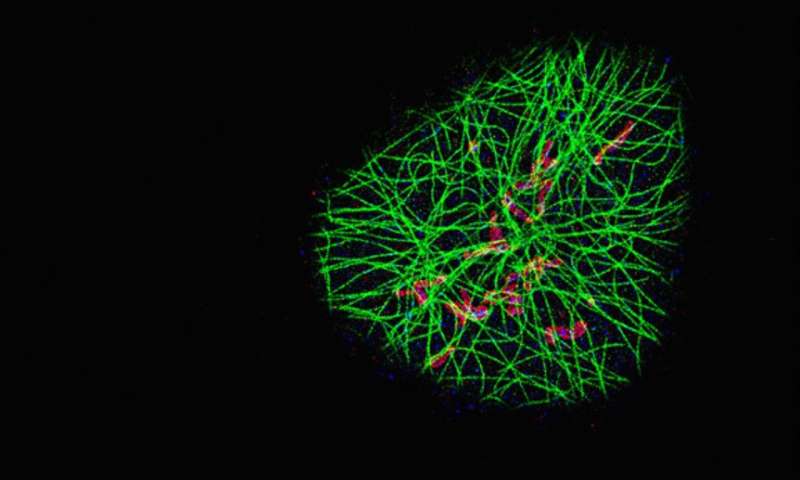
"Our new approach, SDC-PAINT, integrates the versatile super-resolution capabilities of DNA-PAINT with the optical sectioning features of confocal microscopes. We thus created the means to explore the entire depth of a cell, and to visualize the molecules within it at the nanometer scale," said Jungmann. The team mapped out the presence of different combinations of protein within whole cells and then went beyond that. "By diversifying our labeling approaches, we also visualized different types of individual biomolecules in the chromosome-containing nucleus, including sequences in the DNA, proteins bound to DNA or the membrane that encloses the nucleus, as well as nuclear RNAs," adds Yin, who is also co-leader of the Wyss Institute's Molecular Robotics Initiative, and Professor of Systems Biology at Harvard Medical School. .
In principle, confocal microscopes use so-called pinholes to eliminate unwanted out-of-focus fluorescence from image planes above and below the focal plane. By scanning through the sample, plane after plane, researchers can gather the fluorescence signals emitted from molecule-bound dyes over the entire depth. Specifically, the MPI/Wyss Institute team developed the technique for "Spinning Disk Confocal" (SDC) microscopes that detect fluorescence signals from an entire plane all at once by sensing them through a rotating disc with multiple pinholes. Moreover, "to achieve 3D super-resolution, we placed an additional lens in the detection path, which allows us to archive sub-diffraction-limited resolution in the third dimension" said first author Florian Schueder, a Graduate Student working with Jungmann who also worked with Yin's Wyss Institute team as part of his master thesis.
"This addition can be easily customized by manufacturers of SDC microscopes; so we basically implement super-resolution microscopy without complex hardware changes to microscopes that are generally available to cell biologists from all venues of biomedical research. The approach thus has the potential to democratize super-resolution imaging throughout whole cells and tissues," said Jungmann.
"With this important advance, super-resolution microscopy and DNA-PAINT could become more accessible to biomedical researchers, accelerating our insights into the function of individual molecules and the processes they control within cells," said Wyss Institute Founding Director Donald Ingber, M.D., Ph.D., who is also the Judah Folkman Professor of Vascular Biology at HMS and the Vascular Biology Program at Boston Children's Hospital, as well as Professor of Bioengineering at SEAS.
More information: Florian Schueder et al, Multiplexed 3D super-resolution imaging of whole cells using spinning disk confocal microscopy and DNA-PAINT, Nature Communications (2017). DOI: 10.1038/s41467-017-02028-8
Journal information: Nature Communications
Provided by Harvard University