Virtual reality for bacteria
Scientists at the Institute of Science and Technology Austria (IST Austria) have managed to control the behavior of individual bacteria by connecting them to a computer. The interdisciplinary team including experimental biologist Remy Chait and mathematician Jakob Ruess (now at the Institut Pasteur and Inria Saclay in France), as well as Professors Calin Guet and Gasper Tkacik, used the setup to build a genetic circuit that is partly living and partly digital. In the experimental proof of concept, they made gene expression in bacteria oscillate, and controlled the patterns of oscillation by adjusting digital communication between individual bacteria. A potential application of such bio-digital hybrid technology could make it possible to "debug" complex biological systems in the same way complex computer codes are debugged: by testing each part individually while simulating its surroundings in a form of virtual reality.
When synthetic biologists want to engineer a microorganism that can fulfill a certain task as part of its metabolic cycle, such as producing a cancer drug or an antibiotic, they usually have to make a significant number of changes to the original organism. Each of these changes has several effects that could interfere with the effects of all other changes, altering the final result. "Even if you understand what the different parts do, you don't know what happens when you put them together," explains Remy Chait. "There is feedback between them that makes the behavior of the full circuit unpredictable."
A potential solution to this problem comes from software development and is called unit and integration testing. In this approach, each component is tested individually and its interaction with the surroundings is studied. The best way to do this is to simulate the surroundings in a virtual space and to let the component interact with this virtual world. It is this method that the researchers now propose to apply also to biological systems.
"Biological systems are complex, and we would benefit if we could debug them like a computer code. In unit and integration testing, you simulate the environment and plug each of the components in separately to verify that they function as intended. Then you combine them in pairs and start all over. In this way, you will see at which point feedback and interference start to disturb the system, and adjust it appropriately," says Remy Chait. By iterating this method, the virtual part could be steadily reduced until the system is fully biological again, and has the desired function.
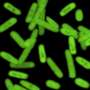
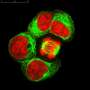
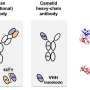
The researchers demonstrated the feasibility of bio-digital hybrids with a bio-digital oscillator. In their setup, modified E.coli cells produce a protein that fluoresces blue-violet. This colored light forms the interface with the digital side. Every six minutes, the computer measures how much light the cell produces, and accumulates a virtual signal molecule in proportion to it. When the signal exceeds a certain threshold, production of the fluorescent protein by the cell is switched off. This is done by a projector that projects red or green light as an "off" or "on" signal onto the light-sensitive cells and thereby links the digital component back to the living parts of the circuit. "The cells are interacting with the simulated environment. What they do influences what the computer does, and what the computer does influences the reaction of the cells. If you know Star Trek, you have certainly heard of the Holodeck. What we have built is essentially a simple Holodeck for genes of microorganisms."
When the researchers tested their hybrid circuits, the population of cells glowed in blue violet—and the glow oscillated, albeit with variations between the individual bacteria. But the researchers wanted the bacteria to oscillate in synchrony, so they altered the digital component and set up a virtual communication network between the bacteria. In this set-up, some of the virtual signal is distributed between neighbors and the group of bacteria display different types of collective oscillation.
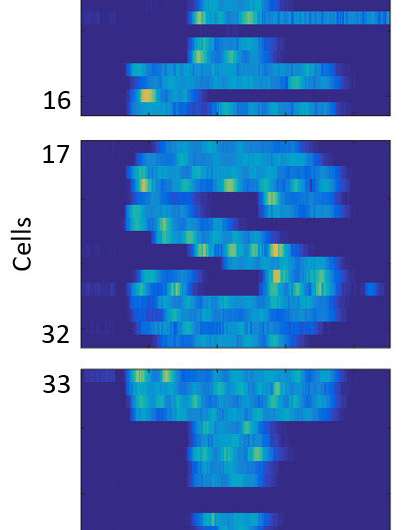
A different application of the researcher's platform is feedback control of individual cells that guides them along pre-specified trajectories of fluorescent gene expression. In this way, they could make a group of cells trace pictures or letters over time (see illustration).
More information: Remy Chait et al, Shaping bacterial population behavior through computer-interfaced control of individual cells, Nature Communications (2017). DOI: 10.1038/s41467-017-01683-1
Journal information: Nature Communications
Provided by Institute of Science and Technology Austria