Nanoscale platform aims to control protein levels
A nanoscale antibody first found in camels combined with a protein-degrading molecule is an effective new platform to control protein levels in cells, according to Rice University scientists. The technique could aid fundamental research into cellular dynamics as well as the design of synthetic gene circuits.
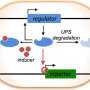
Rice chemical and biomolecular engineer Laura Segatori, former graduate student Wenting Zhao and former undergraduate Lara Pferdehirt invented a bifunctional recognition system they call NanoDeg. It allows them to target specific proteins in a cell and strictly regulate their degradation.
The plug-and-play system will allow synthetic biologists to study the function of a specific protein within the cellular environment by assessing how the protein expression level affects the life of a cell, Segatori said.
The research appears in the American Chemical Society journal ACS Synthetic Biology.
NanoDeg accelerates proteolysis—the enzymatic breakdown of proteins—to control the levels of targeted proteins after translation.
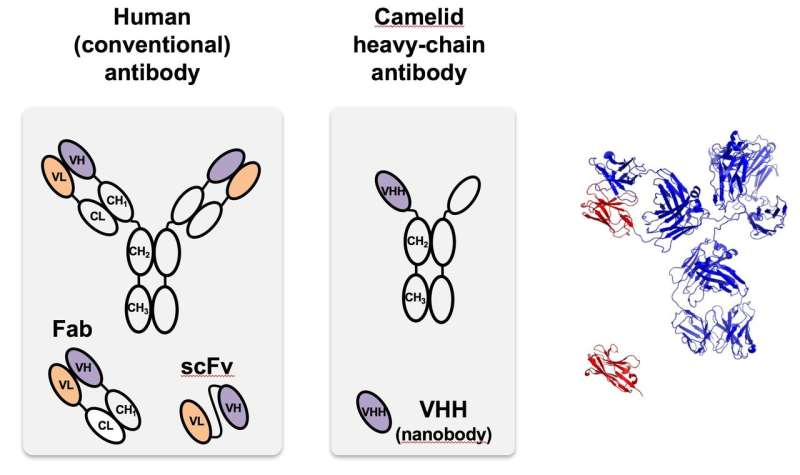
One function springs from the single-chain antibody from camelids, which can be customized to target specific proteins. When the antibodies were discovered in camels (and later sharks), researchers quickly recognized their unique properties, including their small size, high solubility and ability to recognize even targets that are hidden or in intermediate states. They are much smaller than the antibodies found naturally in humans and most other organisms but can be readily made and modified in bacteria and other cells.
The other function relies on degrons, short sequences in proteins that are responsible for regulating the rate of a protein's degradation. These can also be customized to tune the depletion of a target protein to the desired levels.
When combined as NanoDegs, they become a powerful, universal platform for modulating cellular protein levels, Segatori said.
"Essentially, it allows us to control the specific amount of proteins in cells," she said. "We can tailor it to target any protein in a cell, and once the degron-tagged nanobody binds to that partner, the whole complex is degraded.
"The advantage of this system is that it targets expression at the protein level," Segatori said. "Typically, when people want to modulate the amount of proteins in cells, they act at the DNA or RNA—the genetic—level. But by acting at the protein level, we can target different post-regulation modifications, and much more importantly, we have much more control over the rate and extent of depletion of the protein."
As a proof of principle, the researchers designed a synthetic gene circuit that expressed both green fluorescent protein (GFP), which researchers use to report on cellular processes, and a NanoDeg that targets it. "We used GFP because it is a commonly used reporter and fluorescence is easy to measure," Segatori said. "When the nanobody recognizes GFP, the whole complex is taken for degradation."
It will be also useful to those who want cleaner information about the activities of proteins in cells.
"Say you design a genetic circuit in which GFP expression is activated when the cell is under stress, like nutrient starvation or heat, for example," Segatori said. "When the cell is exposed to the stimulus, GFP is expressed and you can detect an increase in cell fluorescence.
"But when you take away the stimulus, the decay of the signal doesn't necessarily reflect the decay of the stimulus; it reflects the stability of the GFP reporter," she said. "What we've done is create a gene circuit in which GFP expression is activated under stimulus, but when the stimulus is turned off, the NanoDeg degrades GFP very rapidly. That increases the sensitivity and dynamic resolution of a synthetic gene circuit."
More information: Wenting Zhao et al, Quantitatively predictable control of cellular protein levels through proteasomal degradation, ACS Synthetic Biology (2017). DOI: 10.1021/acssynbio.7b00325
Journal information: ACS Synthetic Biology
Provided by Rice University