Elastic incoherent neutron scattering at ILL challenge the Lindemann criterion in proteins
Proteins are the nano-machines that Nature uses to perform most of the processes critical for the metabolism in cells. One of the key goals of life and physical sciences revolves around understanding the structural and dynamic properties of the native, transition, intermediate, and denatured states of proteins. The denaturation transition – defined as the transition of proteins from their specific native functional state to the unfolded inoperative state – is of particular interest, as it is defining the boundaries of stability and functionality of the phase diagram of proteins.
Internal subnanosecond timescale motions are also key for protein folding – without these proteins could not even fold in their native structure. Furthermore, they are extremely sensitive to the amount and nature of the solvent surrounding the protein surface i.e. both the amplitude and rate of these dynamics can be greatly reduced when proteins are embedded in sugar-glass matrices.
Although science knows that these fast fluctuations guide protein conformational changes, their role for protein stability and unfolding still remains elusive.
The results of a novel study conducted at the Institut Laue-Langevin (ILL), through a collaboration between the CNRS' Laboratoire de Biochimie Théorique (France), the Universities of Perugia, Pisa and Verona (Italy) and the CNR (Italy), gave a renewed picture of the Lindemann criterion. When conducting elastic neutron scattering experiments, researchers found a common scaling toward a constant value for the local fluctuations of a model protein in different environments, when approaching the unfolding temperature.
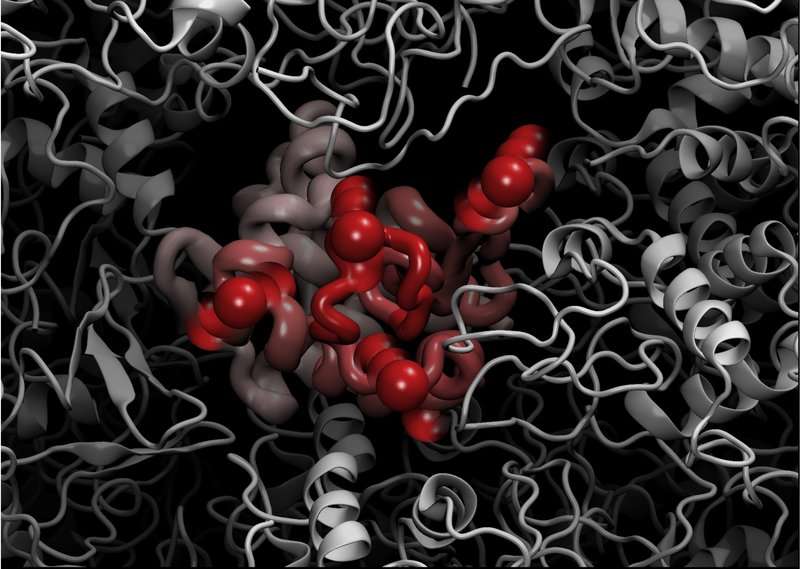
Using the state-of-the-art instruments at ILL, namely the wide-Q range backscattering spectrometer IN13, the researchers conducted elastic incoherent neutron scattering experiments on the lysozyme protein, chicken egg-white lysozyme (CEWL) in the presence of different perdeurated matrices (D20, glycerol, and glucose). This allowed them to study the model protein's sub-nanosecond timescale dynamics in correspondence with the unfolding transition.
This experimental technique is highly sensitive to the motions of hydrogen atoms, and suitable for exploring protein motions on a pico to nano timescale. It yields precise quantitative measurements of the amplitude of protein internal motions in terms of the hydrogen mean-square displacements (MSD).
By combining elastic incoherent neutron scattering and advanced molecular dynamics simulations, they showed that, although different solvents modify the protein melting temperature, a unique dynamical regime is attained when close to thermal unfolding in all solvents tested.
This is reminiscent of the famous Lindemann criterion introduced in 1910, where F.A. Lindemann developed a practical criterion to predict the melting temperature of crystals. Furthermore, the analogy between melting of inorganic crystals and native biomolecules suggests that these seemingly very different systems may share behaviour in corresponding phase transitions.
The common scaling for the protein MSD at melting point not only sheds light on the relationship between protein flexibility and stability, but also opens the possibility to predict protein unfolding in special environments (e.g. the cell interior) by following thermal, local fluctuations.
The criterion they propose can also be applied to investigate the temperature range where micro-organisms thrive e.g. at extreme temperature and pressure conditions in the deep sea or even in space.
This research potentially lays the foundation for a deeper understanding of the folding and unfolding of proteins, which are crucial processes in the metabolism of cells, regulation of biological activity and targeting of proteins to different cellular locations.
Additionally, understanding the functions of protein dynamic is key for biotechnology and pharmaceutical industries, where therapeutic principles based on proteins are worth approximately $30 billion in the US market alone.
More information: Marina Katava et al. Critical structural fluctuations of proteins upon thermal unfolding challenge the Lindemann criterion, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1707357114
Journal information: Proceedings of the National Academy of Sciences
Provided by Institut Laue-Langevin