Spotting the invisible
Chemists at Umeå University have succeeded in mapping structures and functions of a transient enzyme state. By modifying the enzyme adenylate kinase, researchers were able to isolate the molecule and study it using the quantitative techniques X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy. The results have been published in the journal PNAS.
"We have now come one step closer to a general understanding of how enzymes function. This is vital for future designs of novel enzymes in biotechnological applications," says Magnus Wolf-Watz, associate professor at the Department of Chemistry at Umeå University.
Biological life is dependent on a large number of cellular, chemical reactions that are often extremely slow and can take months or years to complete. For chemical and biological time-scales to match, chemical reactions are sped up in cells with the use of enzymes as efficient biocatalysts.
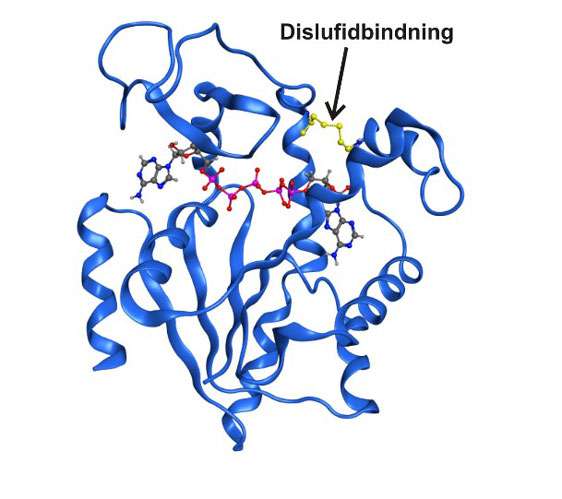
Over the last decade of research it has been made clear that enzyme structures that only exist briefly and transiently can be entirely essential for the catalytic function. So far, it has not been possible to study these states in detail due to the simple fact that they are invisible to traditional spectroscopic techniques. Now, researchers at the Department of Chemistry at Umeå University in Sweden have managed to capture a transient state central to the function in the essential enzyme adenylate kinase. The short-term state was possible to enrich by exchanging two amino acids in the enzyme with the reactive amino acid cysteine.
"It's the first time that anyone has managed to study a transient enzyme state directly using spectroscopic and quantitative techniques. With the use of our methods, we were able to in detail describe both the structure and the function of the enzyme in the specific transient phase," says Magnus Wolf-Watz, who led the study together with postdoc Michael Kovermann, who is now holding a group leadership at the University of Konstanz in Germany.
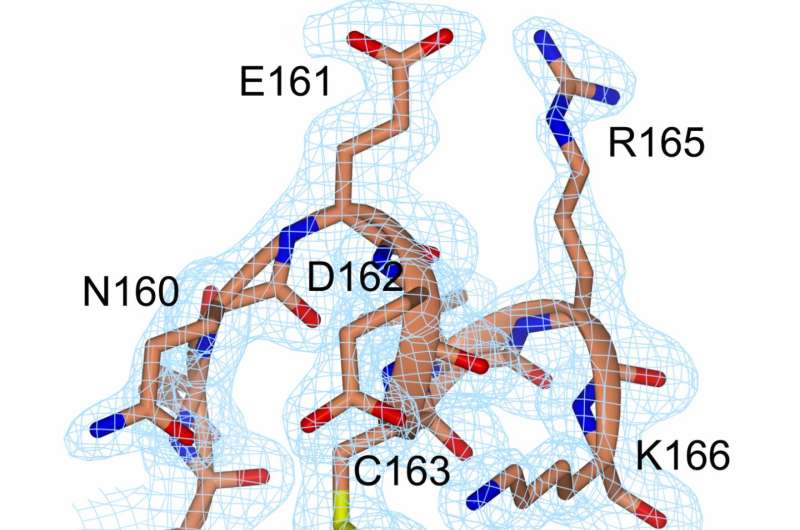
The results indicated that the function in one enzyme is entirely dependent on its inherent dynamics, and without dynamics the enzyme would be rendered useless. Furthermore, it turned out that the transient state bound its substrate molecules much stronger than the natural proteins. The results gave us further clues as to how enzymes can speed up reactions with such incredible specificity and efficacy. All the while, the method that we developed can be generally useful in studies of other enzymes."
Magnus Wolf Watz's NMR team has collaborated with the x-ray crystallographers Uwe H. Sauer and Elisabeth Sauer-Eriksson on the study.
"We have had a long-term and very productive collaboration. This is a good example of how important the collaborative atmosphere found at Umeå University is, and how we should cherish and further develop it in future," says Magnus Wolf-Watz.
More information: Michael Kovermann et al. Structural basis for ligand binding to an enzyme by a conformational selection pathway, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1700919114
Journal information: Proceedings of the National Academy of Sciences
Provided by Umea University