Weather forecasting technology used to predict where proteins anchor within human cells
Met Office technology used to study climate change is being used by scientists to predict the behaviour of vitalsorting and location of proteins cells in cells of the the human body.
Proteins are large biomolecules, which are required for the structure, function, and regulation of the cells in the human body. After their production, many proteins have to be routed to specific locations within the cell for it to work properly.
Researchers at the University of Exeter have made a key finding about the signals and mechanisms which mediate and regulate this protein sorting in the cell. They have discovered how an important group of membrane proteins, which function as adaptor molecules at the surface of cellular compartments, are routed to their specific target membranes.
Proper targeting of these membrane adaptors is crucial for the function of cellular compartments. Mis-targeting or loss of adaptor function leads to a range of severe or fatal disorders associated with metabolic, developmental or neurological defects.
Dr Michael Schrader, from the University of Exeter, who led the study, said: "This research has allowed us to better understand how the cell sorts and delivers these proteins. We now know which properties determine how they travel to specific organelles and can exploit this knowledge to predict the localisation of hitherto uncharacterised adaptor proteins encoded in the human genome."
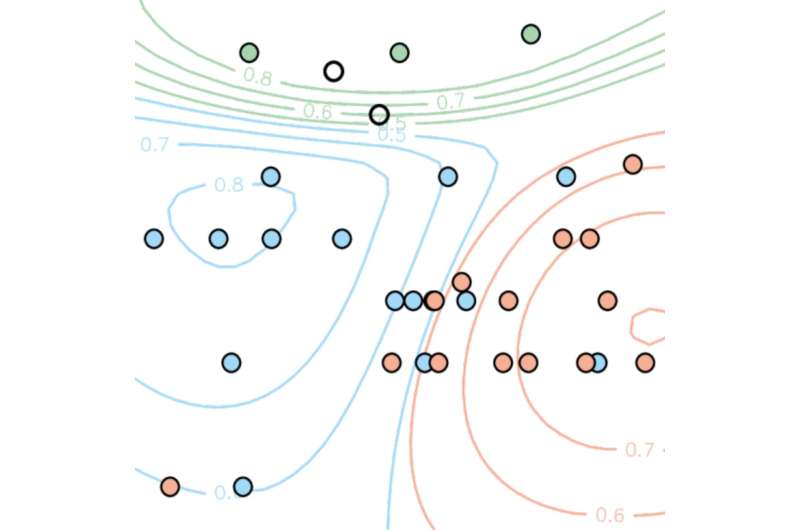
The study has identified for the first time the specific chemical properties in tail-anchored membrane proteins which determine where they travel in a cell. It reveals that a combination of tail charge and hydrophobicity - or water-fearing - of the membrane-spanning domain determines targeting to distinct organelles. Subtle alterations in those physicochemical parameters are sufficient to shift adaptor protein targeting between organelles.
Dr Schrader said: "Our findings can be used to modulate and improve targeting of membrane adaptors. This could also improve medical treatment, because researchers could use this information to produce drugs which better target specific organelles."
The researchers are working with experts from the Met Office. "It can be expensive and slow to run the most accurate computer simulations of the Earth's climate, so we are always looking for ways to make the best use of the data we have. It turns out the statistical methods we use to predict climate change are useful for predicting where proteins anchor within the cell", said co-author Doug McNeall from the Met Office.
More information: Joseph L. Costello et al, Predicting the targeting of tail-anchored proteins to subcellular compartments in mammalian cells, Journal of Cell Science (2017). DOI: 10.1242/jcs.200204
Journal information: Journal of Cell Science
Provided by University of Exeter