Rules devised for building ideal protein molecules from scratch

(Phys.org)—By following certain rules, scientists can prepare architectural plans for building ideal protein molecules not found in the real world. Based on these computer renditions, previously non-existent proteins can be produced from scratch in the lab. The principles to make this happen appear this month in Nature magazine.
The lead authors are Dr. Nobuyasu Koga and Dr. Rie Tatsumi-Koga, a husband-and-wife scientific team in Dr. David Baker's lab at the University of Washington Protein Design Institute.
The project benefited from hundreds of thousands of computer enthusiasts around the world who adopted Rosetta@home for simulating designed proteins.
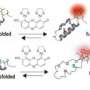
Protein molecules start as an unstable, high energy chain of amino acids. This chain then begins folding into various shapes to try to achieve a stable, low energy state. The end result is its distinctive molecular structure. Rosetta@home volunteers helped the project team to plot this energy landscape from protein structure predictions.
"The structural options become fewer as the interactions that stabilize the protein selectively favor one folding pattern over others," explained Koga. "This decline in conformation options to eventually achieve a unique, ordered structure is called a funnel-shaped energy landscape," he said, drawing a tornado-like figure on a whiteboard. The researchers came up with guidelines for robustly generating this type of energy landscape.
According to Tatsumi-Koga, these rules require the interactions among the residues in the protein's amino acid chain to consistently favor the same folded conformation in forming its molecular shape. This is made possible, for example, by defining whether a specific unit will form a "right-handed" orientation or its mirror image, and disfavor others.
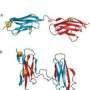
The researchers, she said, synthesized the proteins they had originally designed and tested "in silico" (on the computer) and physically characterized them through "in vitro" (laboratory test tube) experiments. They also compared the molecular structures of the computer models with these laboratory-derived proteins to see how well they matched.
Koga stressed that the project looked strictly at protein structure. He smiled as he said his group was striving toward a "platonic ideal," a reference to Plato's theory of perfect forms. In our imperfect material world, proteins are not always optimized for their stability, but can be beset by bulges, kinks, strains, and improperly buried parts, and many diseases arise from protein malformations.
During this project, the researchers achieved a library of five ideal structures, but since filing their report have added several more. To make them accessible to other scientists, the designs have been deposited in the Research Collaboratory for Structural Bioinformatics and the lab analysis of their chemical structure was put in the Biological Magnetic Resonance Database.
The team was not attempting to create specific new proteins that could carry out particular activities.
However, their design principles and methods, according to their report, should allow the ready creation of a wide range of robust, stable, building blocks for the next generation of engineered functional proteins. Such proteins would be custom-made for the task, instead of repurposed from proteins with unrelated functions.
The hope is that engineered proteins will be useful for drug and vaccine development, especially for formidable viruses like HIV or rapidly changing ones, like the flu. Proteins designed to exact specifications might also prove therapeutically useful in cleaving mutated genes, and for speeding up chemical reactions important in industry and environmental reclamation.
Journal information: Nature
Provided by University of Washington