This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Reimagining the undergraduate bio lab experience, and keeping students in STEMM
HHMI's Science Education Alliance is transforming the undergraduate introductory science experience at more than 150 two- and four-year institutions across the US. These same students are making discoveries that could change how scientists tackle bacterial infections and antibiotic resistance.
First impressions are difficult to shake. That's why the Howard Hughes Medical Institute's Science Education Alliance (SEA) is changing the introductory science experience for thousands of undergraduate students and their faculty while transforming students into researchers early in their academic journey.
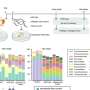
In the Annual Review of Virology HHMI shared a summary of findings about the SEA Program and the impact of course-based research experiences on student persistence in science.
Established in 2008, HHMI's SEA supports a community of faculty from more than 150 two- and four-year undergraduate institutions, including community colleges, tribal colleges, liberal arts colleges, and research universities. The SEA provides faculty at participating institutions with a uniquely designed research curriculum, training, and year-round scientific support, enabling SEA faculty to mentor their students to work on high-impact research projects early in their academic tenure.
SEA students and their faculty pivot away from the typical 100-level science course structure, which all too often centers on lectures and planned lab exercises. Instead, SEA students—typically, first- or second-year undergraduates—venture beyond the classroom to, quite literally, dig into discovery science.
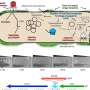
At the heart of the SEA Program is Phage Hunters Advancing Genomic and Evolutionary Science (SEA-PHAGES). SEA-PHAGES is a two-semester, course-based research project aimed at undergraduate students who are new to college-level science and who have little to no research experience.
During their first SEA-PHAGES term, students collect local soil samples to isolate bacteriophages—viruses that infect and kill bacteria. The students then apply a variety of microbiology techniques to purify and characterize their bacteriophages, or "phages" as they are commonly called.
In their second SEA-PHAGES term, students interpret the arrangement and function of the phage's genes through a process known as genome annotation using bioinformatics software. Because phages are remarkably diverse and abundant—there are roughly 1031 phages on Earth—students are all but guaranteed to find a virus or annotate a gene that has never been seen before. Many SEA students celebrate their first scientific discovery, naming their newly discovered phage before they even choose their undergraduate major.
Just like other HHMI scientists, SEA students and instructors meet to present and discuss their research findings at local science meetings. Several thousand researchers attend the annual SEA Symposium each year, making it HHMI's largest science meeting.
To take their findings a step further, students submit their annotated phage genomes to GenBank, the National Institutes of Health genetic sequence database for all publicly available DNA sequences. Many SEA students also work with their faculty and peers to publish their findings as co-authors of scientific papers.
"One of the biggest successes of our SEA-PHAGES Program is the fact that our students are in a much stronger position to compete for opportunities," said Marie Fogarty, a biology faculty member who oversees the SEA-PHAGES Program at Durham Technical Community College in North Carolina.
Fogarty noted that Durham Tech SEA-PHAGES students have secured coveted spots in National Science Foundation Research Experience for Undergraduates summer programs and other programs or paid internship opportunities at Duke University, North Carolina State University, and the University of North Carolina, Chapel Hill (UNC), as well as at institutions as far as Montana State University.
Many students have graduated from Durham Tech and continued in science, technology, engineering, mathematics, or medicine (STEMM) at NC State or UNC. One of Fogarty's recent graduates turned down a full ride at a nearby institution to continue her studies at Duke University. "All of a sudden, the community college is not a 'less than,' it is an 'equal to' experience," added Catherine Ward, Durham Tech's chair of biological sciences.
What is a phage?
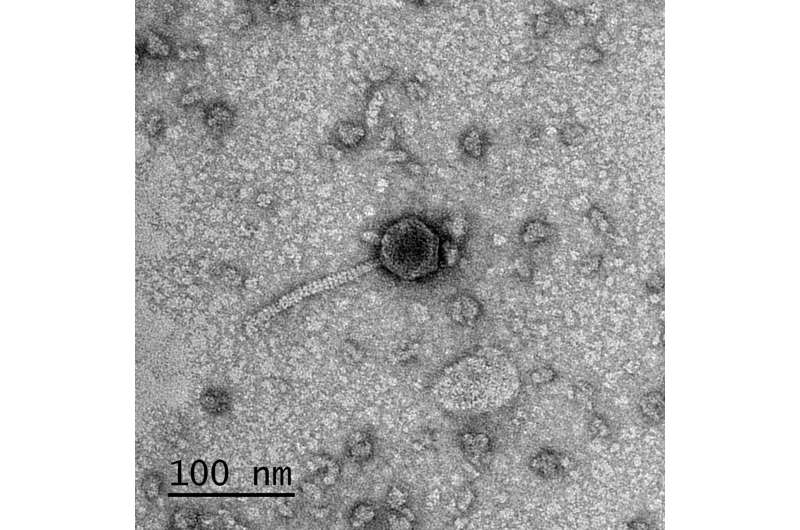
A bacteriophage, commonly referred to as a "phage," is a virus that infects and kills bacteria. There are roughly 1031 phages on Earth, accounting for about 1023 infections of bacteria per second. Virologists have found a tremendous diversity of phages in every ecosystem, from the bottom of the ocean floor to the human gut.
SEA-PHAGES was first established as an expansion of HHMI Professor Graham Hatfull's Phage Hunters Integrating Research and Education (PHIRE) program at the University of Pittsburgh. Working with the Pittsburgh team, Tuajuanda Jordan, now president of St. Mary's College of Maryland, helped to launch SEA-PHAGES with an initial cohort of just a dozen schools. To date, more than 50,000 students have participated in the program, which supports more than 5,500 undergraduates annually.
Each year, the SEA welcomes about a dozen new institutions to the program. Faculty who are interested in launching a SEA program at their institution the following academic year must apply by October 31. SEA Program Officer Pushpa Ramakrishna and Senior Program Assistant Billy Biederman, who lead the SEA's recruitment efforts, emphasize that this is not a typical competitive application process.
"Since the SEA has the potential to revolutionize undergraduate science education, all institutions of higher education are encouraged to apply," Ramakrishna said.
"Although certain elements need to be in place at an institution to enable students to successfully engage in SEA research, we collaborate closely with all interested faculty to make it feasible for them to implement SEA research successfully at their institutions."
Biederman added, "We encourage any faculty that want to do course-based research with their students, even if you've never worked with viruses before, to reach out to us and have a conversation."
Of the SEA-PHAGES institutions, 35 have also established a SEA-GENES program since its launch in 2019. Short for "Gene-function Exploration by a Network of Emerging Scientists," SEA-GENES extends the SEA-PHAGES Program with an additional year of course-based research for mid-level undergraduates, through which students work on building phage gene libraries and screening phages for gene functions.
As with SEA-PHAGES, SEA-GENES allows student scientists to contribute useful resources and experimental observations to the broader scientific community.
Most recently, in May 2024, Richard Pollenz, director of the SEA-PHAGES and SEA-GENES Programs at the University of South Florida (USF), teamed up with 27 USF SEA-GENES students to publish their findings on a bacteriophage named Girr that infects the bacterium Mycobacterium smegmatis, a close relative of human pathogens such as Mycobacterium tuberculosis. The group's paper represented two years of work spanning two SEA-GENES cohorts.
"I am incredibly proud of our recent SEA-GENES publication," Pollenz said, noting that the group's earlier preprint led to queries from eight different scientific journals. "This speaks to the quality and impact of the research. It was amazing how excited the students were to see the final product and appreciate what they had contributed."
Central to the SEA's overall success is a mission to dismantle barriers to scientific research and discovery. The SEA is designed as an inclusive Research-Education Community (iREC), where centralized research support enables faculty from a diversity of colleges and universities to provide their undergraduates with research experience regardless of their institutional research capacities.
While some students who enroll in SEA-PHAGES have an interest in science, others find that the experience opens their eyes to a world they had never considered.
"There's data that show that students' introductory science experiences, whether they are in biology or chemistry, can serve as gatekeepers or weed-out spaces," said Danielle Heller, HHMI's SEA Program Officer and SEA-GENES lead scientist.
"Typically, the way that science is taught in most places is not the way that science is actually done. And so, by incorporating real, authentic research into courses, you're not only giving students a better experience that's more aligned with the reality of doing science and being a scientist, but you're also immediately integrating them into the ecosystem in a way that has important scientific impacts."
For more than 15 years, the SEA Program has shown that engaging undergraduates in discovery science can keep more students in STEMM.
According to a 2017 report from the US National Academies of Sciences, Engineering, and Medicine, students who participate in undergraduate research experiences are more likely to earn a STEMM degree than their peers. Even more, graduates of SEA programs often share how their experiences influenced their decision to stay in the field.
Heller fondly recalls how, a few years ago, she and SEA Senior Program Lead Vic Sivanathan gave a presentation at a large Massachusetts-based biotech company. During the talk, a woman in the audience stood up and declared, "I was a SEA-PHAGES student and that is the reason I'm in this room today."
Across the US, countless students attend institutions—such as community colleges—that may lack the resources needed to support hands-on research experiences. According to the Community College Research Center housed at Columbia University, community colleges enroll approximately 40% of all undergraduate students in the US and serve disproportionately high numbers of women, Black, Indigenous, and Latine students, as well as low-income students, first-generation college students, and students with disabilities.
"It's pretty amazing that our [SEA] students have the opportunity to do the kind of research we're doing at our tiny rural community college," said Matthew Fisher, a biology professor and head of the SEA program at Oregon Coast Community College.
In a typical introductory biology course, students learn some of the fundamentals mostly by reading from a textbook and completing basic lab assignments, but Fisher said he wanted more for his students. "The reason why I applied to the SEA-PHAGES Program is because I wanted students to immerse themselves in the scientific process. I didn't want to just perpetuate this notion that when you take a science class it's just about memorizing facts."
Since 2008, SEA undergraduate researchers have isolated more than 23,000 phages, of which more than 4,500 are fully sequenced and annotated. Further, SEA students have functionally characterized hundreds of genes, thereby shedding new light on how phages could be used as therapeutics to treat bacterial infections, including those resistant to antibiotics.
Why phages?
All told, phages carry out roughly 1023 infections of bacteria per second. Virologists have found phages in every ecosystem ranging from the ocean floor to the human gut, and the morphologies and genomic content of phages can vary wildly.
"Part of the outcome of the SEA-PHAGES Program has been large numbers of completely sequenced phage genomes spanning this great diversity," Hatfull said.
"Each of those phages has an average of 100 genes or so, and we don't know what most of them do. The question of 'What do these genes do?' is relevant to virtually everything. It's a fundamental, compelling mystery in science, and it represents a critically important question for understanding how phages could be used therapeutically."
Although countless phage genes remain a mystery, advances in protein structure prediction programs—like AI-powered AlphaFold—enable SEA faculty and students to "make sense" of phage genome data, Hatfull said.
"Advances in computational programs enable scientists to make predictions of how proteins may interact, and that will lead to hypotheses that can be tested experimentally to allow us to see what various phage genes are doing. I see it as a game-changer."
On the education front, phages offer a beautiful entry point for discovery science. Students and faculty collaborate to identify new phages, and at times they may need to work together to overcome roadblocks. Many SEA faculty enter the program having no prior experience with phages or even virology. The SEA provides hands-on training, course materials, bioinformatics software, and a support community to help faculty excel.
Hatfull points out that the wider scientific community has a lot to learn about phages, and student-driven discoveries are making an impact. In 2019, a team of scientists showed that an experimental, personalized treatment using genetically engineering bacteriophages combatted a Mycobacterium infection in a 15-year-old girl with cystic fibrosis. To do it, they called Hatfull for help.
Because of the SEA Program, Hatfull oversees the world's largest collection of phages. He and his team combed through any that might fight the bacteria wreaking havoc on the patient's immune system.
They zeroed in on three phages—named Muddy, ZoeJ, and BPs—which offered promising characteristics. Hatfull and his colleagues modified the genomes of two and combined them with the third to create a "phage cocktail." After doctors gave the patient an IV of the cocktail twice each day for six weeks, the patient's infection had almost completely disappeared.
More broadly, phage-based therapies could provide a new weapon against an increasingly dire public health challenge: drug-resistant bacteria. According to the UN Environment Program, in 2019, nearly 1.3 million deaths worldwide were directly attributed to drug-resistant infections.
"The major goals of the SEA-PHAGES and SEA-GENES Programs are to understand viral diversity, evolution, and origins," Hatfull said.
"The discovery of so many phages and their genomic characterizations are an incredibly important part of being able to respond to the call for phages to be used therapeutically. Through the SEA Program, students and faculty provide many of the insights that make it possible to explore phage-based therapies."
"So many students enter college wanting to make a difference for humanity," Heller said.
"I think the way we teach science can connect—or disconnect—them from that motivation. Through the SEA Program, students are doing science, and they can see how it connects to the clinic, making a difference for humanity. That's really powerful and motivating."
More information: Danielle M. Heller et al, SEA-PHAGES and SEA-GENES: Advancing Virology and Science Education, Annual Review of Virology (2024). DOI: 10.1146/annurev-virology-113023-110757
Provided by Howard Hughes Medical Institute