This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Lab shows phage attacks in new light
As antibacterial resistance continues to render obsolete the use of some antibiotics, some have turned to bacteria-killing viruses to treat acute infections as well as some chronic illnesses.
Graham Hatfull, the Eberly Family Professor of Biotechnology in the Kenneth P. Dietrich School of Arts and Sciences at Pitt, has pioneered the use of these viruses—bacteriophages, phages for short—to treat infections in chronic diseases such as cystic fibrosis. Although the importance of resistance may have eluded the early discovers of antibiotics, Hatfull is intent on understanding how bacteria become resistant to phages.
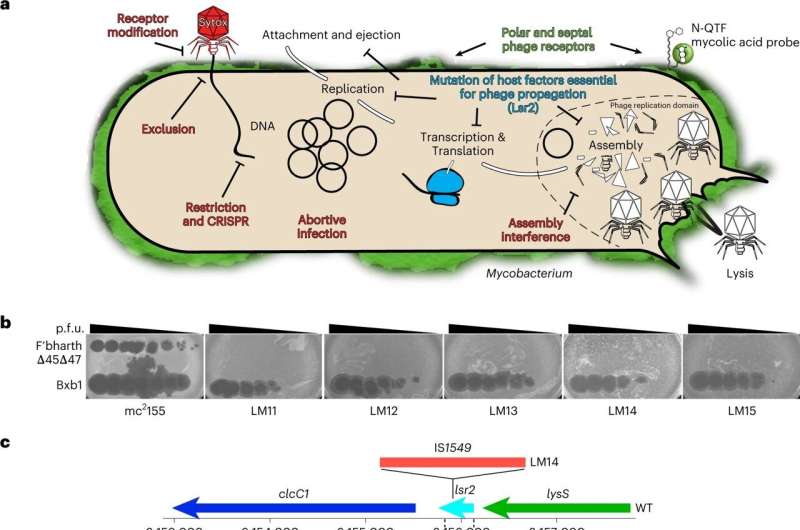
His lab has just discovered how a specific mutation in a bacterium results in phage resistance. The results were published Feb. 23, in the journal Nature Microbiology.
The new methodology and tools his team developed also gave them the opportunity to watch in unprecedented detail as a phage attacks a bacterium. As the use of phage therapy expands, these tools can help others better understand how different mutations protect bacteria against invasion by their phages.
For this study, the team started with Mycobacterium smegmatis, a harmless relative of the bacteria responsible for tuberculosis, leprosy and other hard-to-treat, chronic diseases. They then isolated a mutant form of the bacterium that is resistant to infection by a phage called Fionnbharth.
To understand how the specific mutation in the lsr2 gene helps these resistant bacteria fight off a phage, the team first needed to understand how phages killed a bacteria without the relevant mutation.
Carlos Guerrero-Bustamante, a fourth-year graduate student in Hatfull's lab, genetically engineered two special kinds of phages for this study. Some produced red fluorescence when they entered a bacterial cell. Others had segments of DNA that would stick to fluorescent molecules so phage DNA would light up in an infected cell.
Following the fluorescent beacons, "We could see where the phage DNA entered the cell," Guerrero-Bustamante said. The imaging methods they used were designed by Charles Dulberger, a collaborator and co-first author of the paper who was then at Harvard T.H. Chan School of Public Health.
"We saw for the first time how the phages take that first step of binding to cells and injecting their DNA into the bacteria," said Hatfull, who is also a Howard Hughes Medical Institute Professor. "Then we applied those insights to ask, 'So, how's it different if we get rid of the Lsr2 protein?'"
The link between Lsr2 and phage resistance has not been previously known, but with their new methods and tools, the team clearly saw the critical role it played.
Typically, Lsr2 helps bacteria replicate its own DNA. When a phage attacks, however, the virus co-opts the protein, using it to replicate phage DNA and overwhelm the bacteria. When the lsr2 gene is missing or defective—as in the phage-resistant Mycobacterium smegmatis—the bacteria doesn't make the protein and phages don't replicate enough to take over the bacterial cell.
This was a surprise.
"We didn't know Lsr2 had anything to do with bacteriophages," Hatfull said.
These new tools can be used to uncover all manner of surprises written in the genes of phage-resistant bacteria. It may also help today's researchers and tomorrow's clinicians to better understand and take advantage of phages' abilities while avoiding the missteps that led to antibiotic resistance.
"This paper focuses on just one bacterial protein," and its resistance to just one phage, Hatfull said, but its implications are wide. "There are lots of different phages and lots of other proteins."
More information: Charles L. Dulberger et al, Mycobacterial nucleoid-associated protein Lsr2 is required for productive mycobacteriophage infection, Nature Microbiology (2023). DOI: 10.1038/s41564-023-01333-x
Journal information: Nature Microbiology
Provided by University of Pittsburgh