This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Researchers directly detect interactions between viruses and their bacterial hosts in soil
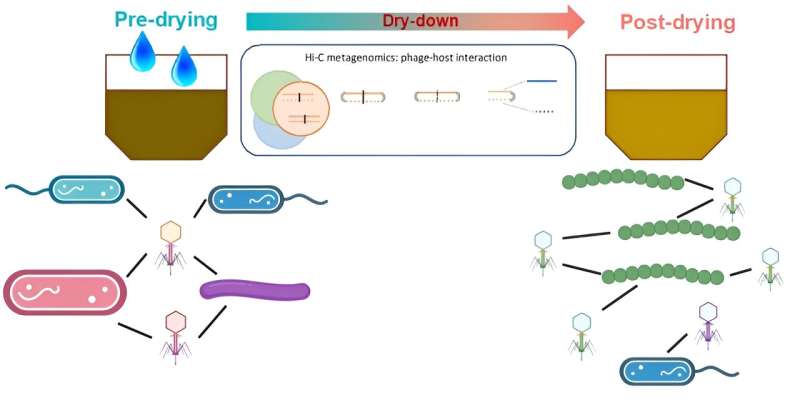
Bacteriophages—viruses that infect bacteria—are common in soil ecosystems. However, many of these phages have not been identified, and the bacteria they target are also a mystery.
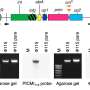
In a study, published in Nature Communications, researchers employed an advanced technology called high-throughput chromosome conformation capture (Hi-C) to directly investigate the relationships between phages and their bacterial hosts in soils with varying moisture levels.
The research found that phages are less active in dry soil than in wet soil. These phages targeted bacterial hosts that are able to survive and thrive in dry conditions. Some of these bacterial hosts appeared to be important species in their bacterial communities. This shows that phages can influence bacterial populations in response to changes in soil moisture.
This study is a significant advance in the use of Hi-C sequencing in soil environments. This technology allows researchers to observe and record the ongoing viral infections and identify the interactions between viruses and their hosts.
Notably, this offers experimental evidence of viral generalists. Previously, this concept had only been predicted based on data analysis. Additionally, by combining Hi-C with other DNA and RNA sequencing techniques, scientists can gain new perspectives on how soil drying influences the relationships between phages and their hosts.
The researchers collected soil samples before and after a two-week incubation period, which simulated the natural drying process that occurs in the field.
To identify the interactions between phages and their hosts, the researchers employed Hi-C, which involves chemically linking the DNA molecules of phages and their bacterial hosts that are in the same cell. The research also used paired metagenomes, and metatranscriptomes generated by the Joint Genome Institute, a Department of Energy user facility, to examine the composition of phage and bacterial communities and their potential activities.
The investigation revealed that some of the key species within the bacterial community could also serve as hosts for phages.
The researchers observed that the frequency of phage infections per host population had a significant negative correlation with the abundance of the hosts in the soil prior to the drying process. However, after the soil had dried, the phages were less active, but they seemed to selectively target hosts with advantageous traits.
Collectively, these findings provide evidence of how phages can influence the dynamics of bacterial populations in response to changing levels of soil moisture.
More information: Ruonan Wu et al, Hi-C metagenome sequencing reveals soil phage–host interactions, Nature Communications (2023). DOI: 10.1038/s41467-023-42967-z
Provided by US Department of Energy