Turning the volume of gene expression up and down
Gene expression can be turned on and off like a switch, or it can be finely adjusted , as with a volume control knob. Dr Garth Ilsley, research scientist in Prof. Nick Luscombe's unit at the Okinawa Institute of Science and Technology Graduate University (OIST), has developed a mathematical model that shows how to predictably tune gene expression. This was validated experimentally using a technique for adjusting gene expression in fruit fly embryos pioneered by Dr Justin Crocker in the group of Dr David Stern at Janelia Research Campus in the U.S. This study, published in Nature Genetics, has important implications in cellular and developmental biology, with potential applications in stem cell reprogramming and regenerative medicine.
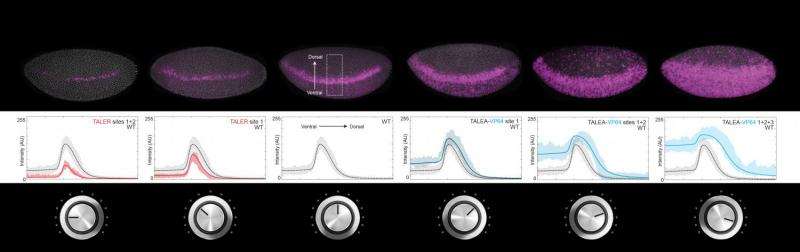
Transcription factors are proteins that bind to special regions of DNA called enhancers, so as to regulate gene expression. Some transcription factors activate gene expression, while others repress it. Gene expression level is like the volume of a radio; some transcription factors turn the volume up, while others turn it down. Scientists have investigated how activation and repression work and how to predict the level of gene expression. In the same way, by turning the volume knob of a radio, it is possible to know how loud or soft the music will be and to regulate it—except that, in this case, each transcription factor has its own volume knob all acting on the same speaker. The challenge is to understand how they all work together to produce the right volume.
Dr Ilsley applied a new mathematical model that does not require information about the number and position of transcription factors binding to the enhancer, which would be like knowing the inner workings of the radio. Instead, the model correctly predicts the final volume only by knowing how the volume knobs are turned. These predictions were tested experimentally using artificial transcription factors that activate and repress gene expression with different strengths.
Scientists worked with fruit fly (Drosophila melanogaster) early stage embryos. The mathematical model shows that expression of genes that determine segmentation of the fruit fly body from head to tail is tunable. Experimental results match the model's prediction, showing that artificial activators and repressors can increase and decrease gene expression gradually in a way that is controllable and reproducible. This is like attaching yet another volume knob to the radio and finding that it works in concert with the existing knobs. Beyond gene expression level, the model was also able to predict in which location in the embryo, for example ventral or dorsal, the gene would be expressed. "It was our dream to bring model and experiment together," enthuses Dr Ilsley.
This study also shows that enhancers can acquire new activators and repressors quite flexibly. "You can bring in foreign transcription factors and the enhancers still work. The enhancers we looked at are not brittle at all. This is evolutionarily important, because it shows how enhancer activity can be adjusted gradually and remain working in changing contexts," points out Dr Ilsley. "Each activator and repressor is like a generic component that takes part in the overall tuning of gene expression. Many possible combinations of natural or artificially engineered transcription factors can produce identical enhancer activities," explains Dr Ilsley.
"We are moving away from having to use an on/off model of gene expression to understand how cell types are specified. Advances in quantitative biology at the single-cell level, like quantitative imaging and RNA sequencing, together with mathematical models, now give biologists the tools they need to delve into the intricacies of gene expression tuning and to predictably manipulate the cell," concludes Dr Ilsley.
More information: Quantitatively predictable control of Drosophila transcriptional enhancers in vivo with engineered transcription factors, Nature Genetics, DOI: 10.1038/ng.3509
Journal information: Nature Genetics
Provided by Okinawa Institute of Science and Technology

















