Scientists capture most detailed images yet of humans' tiny cellular machines
A grandfather clock is, on its surface, a simple yet elegant machine. Tall and stately, its job is to steadily tick away the time. But a look inside reveals a much more intricate dance of parts, from precisely-fitted gears to cable-embraced pulleys and bobbing levers.
Like exploring the inner workings of a clock, a team of University of Wisconsin-Madison researchers is digging into the inner workings of the tiny cellular machines called spliceosomes, which help make all of the proteins our bodies need to function. In a recent study published in the journal Nature Structural and Molecular Biology, UW-Madison's David Brow, Samuel Butcher and colleagues have captured images of this machine, revealing details never seen before.
In their study, they reveal parts of the spliceosome—built from RNA and protein—at a greater resolution than has ever been achieved, gaining valuable insight into how the complex works and also how old its parts may be.
By better understanding the normal processes that make our cells tick, this information could some day act as a blueprint for when things go wrong. Cells are the basic units of all the tissues in our bodies, from our hearts to our brains to our skin and lungs.
It may also help other scientists studying similar cellular machinery and, moreover, it provides a glimpse back in evolutionary time, showing a closer link between proteins and RNA, DNA's older cousin, than was once believed.
"It gives us a much better idea of how RNA and proteins interact than ever before," says Brow, a UW-Madison professor of biomolecular chemistry.
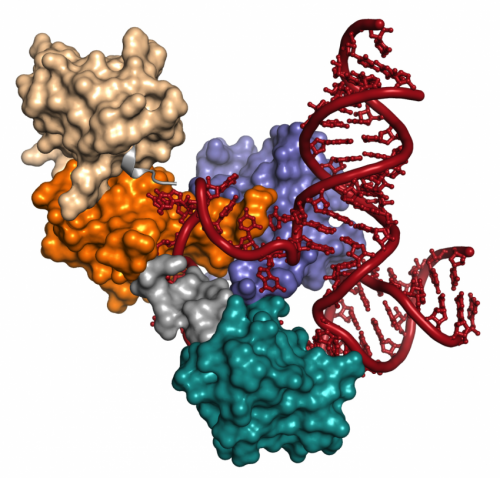
The spliceosome is composed of six complexes that work together to edit the raw messages that come from genes, cutting out (hence, splicing) unneeded parts of the message. Ultimately, these messages are translated into proteins, which do the work of cells. The team created crystals of a part of the spliceosome called U6, made of RNA and two proteins, including one called Prp24.
Crystals are packed forms of a structure that allow scientists to capture three-dimensional images of the atoms and molecules within it. The crystals were so complete, and the resolution of the images so high, the scientists were able to see crucial details that otherwise would have been missed.
The team found that in U6, the Prp24 protein and RNA—like two partners holding hands—are intimately linked together in a type of molecular symbiosis. The structure yields clues about the relationship and the relative ages of RNA and proteins, once thought to be much wider apart on an evolutionary time scale.
"What's so cool is the degree of co-evolution of RNA and protein," Brow says. "It's obvious RNA and protein had to be pretty close friends already to evolve like this."
The images revealed that a part of Prp24 dives through a small loop in the U6 RNA, a finding that represents a major milestone on Brow and Butcher's quest to determine how U6's protein and RNA work together. It also confirms other findings Brow has made over the last two decades.
"No one has ever seen that before and the only way it can happen is for the RNA to open up, allow the protein to pass through, and then close again," says Butcher, a UW-Madison professor of biochemistry.
Ultimately, Butcher says they want to understand what the entire spliceosome looks like, how the machines get built in cells and how they work.
While this is the first protein-RNA link like this seen, Brow doesn't believe it is unique. Once more complete, high-resolution images are captured of other RNA-protein machines and their components, he thinks these connections will appear more commonly.
He hopes the findings mark a transition in the journey to understand these cellular workhorses.
"It's exciting studying these machines," he says. "There are only three big RNA machines. Ours evolved 2 billion years ago. But once it's figured out, it's done."
The U6 crystal structure was imaged using the U.S. Department of Energy Office of Science's Advanced Photon Source at Argonne National Laboratory. The work was funded by a joint grant from the National Institutes of Health shared by Brow and Butcher.
More information: Animation, study paper: www.nature.com/nsmb/journal/va … b/nsmb.2832_SV1.html
Journal information: Nature Structural and Molecular Biology
Provided by University of Wisconsin-Madison