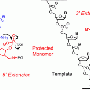
A pathway to bypass DNA lesions in the replication process is experimentally shown
DNA lesions are really common —about one million individual molecular lesions per cell per day— because its long strands usually have one missing base or are damaged. These lesions can stall the DNA replication process, what can lead to the cell death. To avoid it, there are several pathways to bypass lesions in order to continue with the process of DNA replication. One of these processes has been entirely reproduced in vitro using some techniques of manipulation of single-molecules in a study published today in Science, led by the researcher of the University of Barcelona Maria Mañosas.
"This pathway was proposed in the seventies and now we have been able to prove it on a bacteriophage through the manipulation of single-molecules that, oppositely to the traditional biochemical techniques that work with a great number of molecules, allows to study how a protein works on a molecule in real time", explains Mañosas, professor at the Department of Fundamental Physics of the UB, affiliated with the campus of International excellence, BKC.
To study a single-molecule, we used magnetic tweezers, a technique which consists on tethering a DNA hairpin between a glass surface and a magnetic bead. A magnetic system generates a magnetic field which allows manipulating the beads and generates magnetic forces. This system can be used in order to measure the extension changes of DNA strands through the screening of the magnetic beads. According to Mañosas, "proteins' activity over DNA can be inferred from the changes in the extension of the molecule. The changes are due to the proteins' work".
The template switching strategy
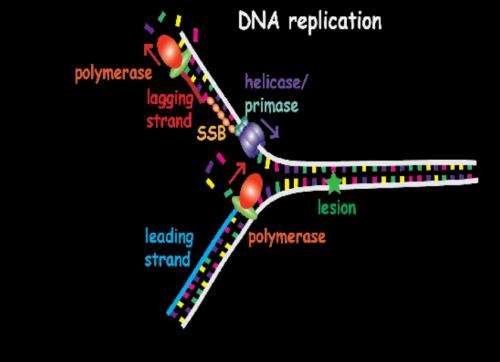
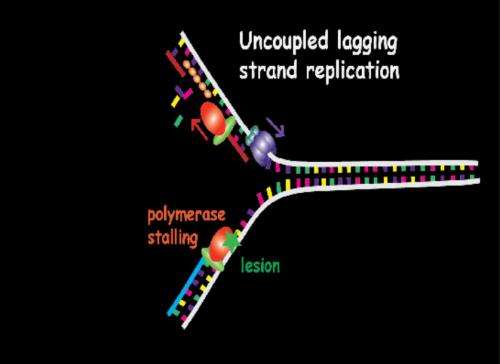
In the DNA replication process, the two strands who act as a template to synthesise a complementary strand are separated, and the new complementary strand joins each of the initial strands in order to obtain two identical copies of the original DNA molecule. In this process take part the polymerases, a family of enzymes that carry out all forms of DNA replication. When in any of the two derived strands there is a lesion, especially in the leading strand, the polymerase stops synthetizing the bases, so the replication process is stalled. "To stall this process can entail some problems in cellular growth", explains Mañosas. "When the replication mechanism (replisome) is disassembled, the bypass process analysed in this study starts", points out the author, member of the Biomedical Research Networking center in Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN) and researcher at the University of Paris.
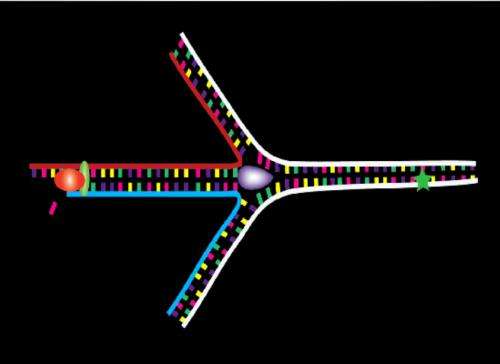
The studied process begins with the action of a helicase protein (UvsW) which promotes the binding of DNA strands, a phenomenon named DNA hybridization. This protein is also able to build an intermediate structure (Holliday junction) taking as a model the not damaged replicated strand and, together with the action of polymerase, drive the system to its departure point, once "jumped" the lesion, and then restart the DNA replication process. "Therefore, the information lost when one strand is damaged can be recovered from the other intact strand which acts as a backup; this process is named "the template switching strategy". In the study, we have also observed the regulation mechanisms of this pathway, as well as the rate of annealing of helicase UvsW, 1500 bases per second, one the largest known", concludes Mañosas.
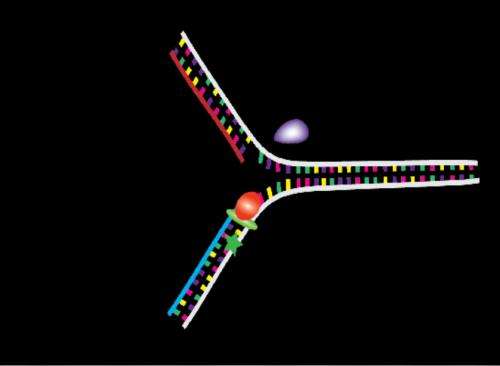
DNA repair is essential in a great number of diseases. A deeper knowledge of these phenomena will enable us to act over some proteins which have similar functions in humans. Mañosas is working on this direction; she is carrying out a study on a human protein named HARP in order to know how it works, because it is known that it has a really important role in the genome conservation and its dysfunction is related to some types of cancer.
More information: Manosas, M., et al., Direct Observation of Stalled Fork Restart via Fork Regression in the T4 Replication System. Science, 30 November 2012: 1217-1220. DOI:10.1126/science.1225437.
Journal information: Science
Provided by Universitat de Barcelona