Researchers study the structure of drug resistance in tuberculosis
(Phys.org) -- Edward Yu took note of the facts – nearly 2 million deaths each year, 9 million infected each year, developments of multidrug-resistant, extensively drug-resistant and now totally drug-resistant strains – and decided to shift his research focus to tuberculosis.
Yu, an Iowa State University and Ames Laboratory researcher, has described in the journal Nature the three-part structure that allows E. coli bacteria to pump out toxins and resist antibiotics.
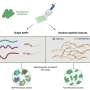
And now, in a paper published online by the journal Nucleic Acids Research, a research team led by Yu describes the structure of a regulator that controls the expression of the multidrug efflux pump in Mycobacterium tuberculosis.
Yu – a professor of physics and astronomy, of chemistry, of biochemistry, biophysics and molecular biology in Iowa State’s College of Liberal Arts and Sciences and an associate of the U.S. Department of Energy's Ames Laboratory – said the latest study is a starting point for a better understanding of how the tuberculosis bacterium is able to resist drugs.
The development of strains totally resistant to drugs “inspired us to move in this direction and try to understand the mechanism in developing drug resistance,” Yu said.
“It is obvious that the emergence of these drug-resistant TB strains has evolved into a major threat and challenges our global prospects for TB control,” Yu’s research team wrote in its latest paper. “Thus, knowledge of the molecular mechanisms underlying drug resistance in M. tuberculosis is essential for the development of new strategies to combat this disease.”
Yu’s research is currently supported by the National Institutes of Health. The researchers’ use of the Advanced Photon Source at Argonne National Laboratory in Argonne, Ill., was supported by the U.S. Department of Energy’s Office of Basic Energy Sciences.
In addition to Yu, the research team includes Qijing Zhang, Iowa State’s Frank Ramsey Endowed Professor of Veterinary Microbiology and Preventive Medicine and the College of Veterinary Medicine’s officer of graduate education; Kanagalaghatta Rajashankar, a senior research associate in chemistry and chemical biology at Cornell University in Ithaca, N.Y., and associate director of the Northeastern Collaborative Access Team facility at the Advanced Photon Source; Iowa State post-doctoral research associates and Ames Lab associates Feng Long and Chih-Chia Su; Iowa State post-doctoral research associate Lei Dai; Iowa State graduate students and Ames Lab student associates Jani Reddy Bolla, Sylvia Do and Hsiang-Ting Lei; recent Iowa State graduate Xiao Chen; and Ames Lab undergraduate summer interns Jillian Gerkey and Daniel Murphy.
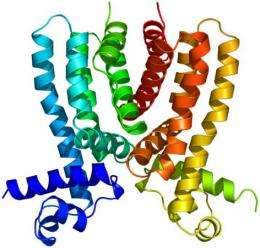
Prior to Yu’s study, not much was known about the structure and function of the tuberculosis efflux pump regulator known as Rv3066.
That, in part, is because researchers have attributed drug resistance in tuberculosis to the bacterium’s very thick cell wall. That wall makes it very difficult to get drugs into the bacterium.
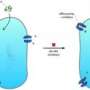
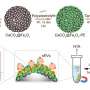
The researchers used X-ray crystallography (including X-ray beams produced by the Advanced Photon Source) to study the Rv3066 structure. They collected data showing the regulator when the toxic compound ethidium bromide was present and when it was not.
The data revealed an asymmetric, two-part molecule with a spiral structure. The structure is flexible, allowing the regulator to recognize and respond to multiple drugs. In the presence of ethidium, Yu’s group says the regulator responds with a rotational motion, inducing expression of the efflux pump that rids the bacterium of antimicrobial drugs.
Studying that structure and mechanism could make a difference in the fight against drug-resistant tuberculosis: “Elucidating the regulatory systems of multidrug efflux pumps in M. tuberculosis,” Yu and the researchers wrote in their paper, “should allow us to understand how this bacterium contributes to multidrug resistance and how it adapts to environmental changes.”
More information: nar.oxfordjournals.org/content … 012/07/19/nar.gks677
Journal information: Nucleic Acids Research
Provided by University of Iowa