This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists discover novel receptor recognition mechanism for alphavirus
Eastern Equine Encephalitis virus (EEEV), an alphavirus, can cause central nervous system infections that can lead to severe encephalitis with a mortality rate of over 30%.
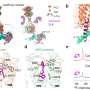
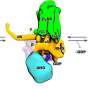
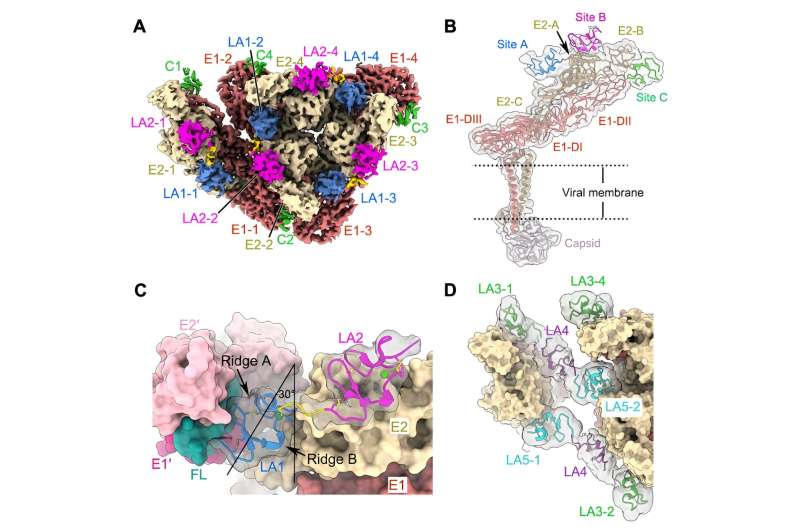
A recent study analyzed multiple high-resolution complex structures of the complete EEEV virion-like particle and the full-length and truncated forms of the LDLR class A repeats (LAs) in the extracellular region of the human very low-density lipoprotein receptor (VLDLR), and identified two main receptor sequential binding modes, LA1-2 and LA3-5.
This discovery updates the understanding of the interaction between alphaviruses and receptors.
The research results, jointly completed by scientists from the Institute of Biophysics (IBP) of the Chinese Academy of Sciences and Tsinghua University, were published in the journal Nature Communications on Aug 10.
VLDLR is a novel alphavirus receptor that binds to the glycoprotein on the alphavirus surface through LAs, its extracellular cysteine-rich repeat sequence domain. However, the specific binding mode and working mechanism remain unclear.
In this study, researchers found three distinct receptor binding sites on the surface of the alphavirus EEEV, all of which are located on protrusions of the E1/E2 glycoprotein trimer far from the membrane surface. The LA domain binding of VLDLR at these three receptor binding sites is not completely fixed.
Through biochemical and cellular experiments, researchers identified the two main receptor sequential binding modes, LA1-2 and LA3-5. Both modes mediate the binding between the virus and cell surface receptors as well as subsequent virus internalization.
Interestingly, the key amino acid residue K206 at site C is not conserved in the EEEV virus. However, the vaccine strain EEEV-PE6 still contains K206 at site C, suggesting a potential risk of using EEEV PE6 strain as a vaccine strain.
The researchers further discovered that a mutation of the key binding site (W50A) in LA1 significantly reduced the virus's ability to adsorb on the cell surface. In contrast, a mutation of the key binding site (W132A) in LA3 enhances the virus's adsorption on the cell surface.
Structural and functional analysis revealed that LA3 and LA1 may competitively bind at site A, and the W132A mutation drives the switch from the LA3-5 dominated binding mode to the LA1-2 dominated binding mode. Thus, the introduction of the dysfunctional LA3 in VLDLR significantly enhances the attachment of EEEV to the cell.
Notably, the VLDLR-W132G mutation was identified in human genome and SNP sequences, indicating that individuals with the W132G mutation may be more susceptible to infection by the EEEV alphavirus.
This study systematically demonstrated the complex and varied receptor binding modes of the novel receptor VLDLR on the surface of the alphavirus EEEV. The work revealed different receptor recognition patterns of the novel receptors in various alphaviruses and improved our understanding on the transmission mechanisms of alphaviruses' extensive invasion across multiple species.
More information: Duanfang Cao et al, The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes, Nature Communications (2024). DOI: 10.1038/s41467-024-51293-x
Journal information: Nature Communications
Provided by Chinese Academy of Sciences