This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers identify structural characteristics of newly emerged SARS-CoV-2 variants
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to adapt to the herd immunity background and evolve into numerous sub-variants.
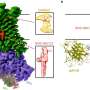
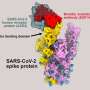
The lab of Prof. George Fu Gao at the Institute of Microbiology of the Chinese Academy of Sciences (IMCAS) reveals the spike (S) protein structures of the recently emerged BA.2.86, JN.1, EG.5, EG.5.1 and HV.1 variants of SARS-CoV-2, and conducted systematic comparative analysis on these sub-variants. The findings are published in Structure.
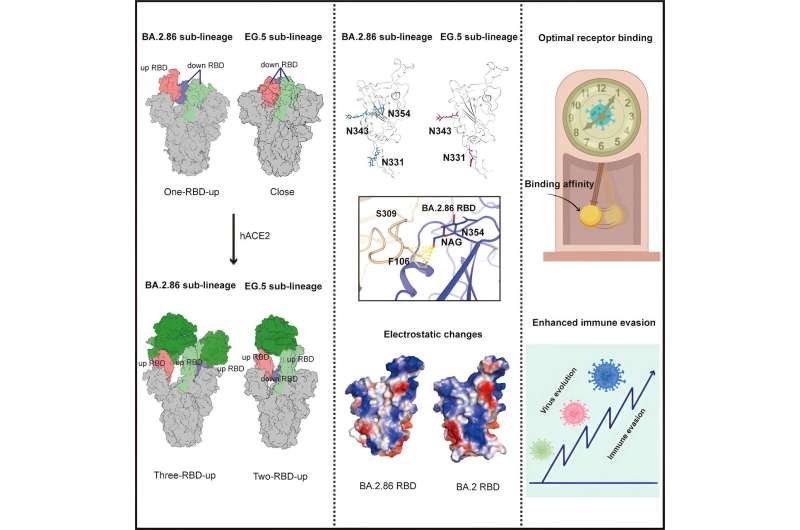
The variants evolved from BA.2 were found to mainly separate into two branches. The branch represented by BA.2.86 and JN.1 was more likely to show the three receptor-binding domains (RBDs)-up state of the S protein under the induction of its receptor, human angiotensin converting enzyme 2 (hACE2), and the branch represented by XBB, EG.5, HV.1 was more likely to show the 2-RBDs-up, 1-RBD-down state under the induction of hACE2.
Compared to other variants, the branch represented by BA.2.86 carried a new N-glycosylation at residue 354 due to the K356T mutation, and this glycosylation site affected the binding to antibodies, such as S309.
Compared with the BA.2 strain circulating in 2022, the RBD surface of the BA.2.86 and JN.1 showed significant electrostatic changes, which enhanced their immune escape. Among them, BA.2.86 had a higher receptor binding affinity than its progeny, JN.1, which escaped from more antibodies.
Structural analysis showed that the R493Q reverting mutation in the BA.2.86 RBD promoted receptor binding, whereas the L455S mutation in the JN.1 RBD reduced the receptor-binding affinity.
This study supports the lab's previous opinion that during the evolution of SARS-CoV-2 variants, immune escape is gradually enhanced, while receptor binding capacity fluctuates and remains in a moderate range (2 digital nanomoles).
More information: Linjie Li et al, Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants, Structure (2024). DOI: 10.1016/j.str.2024.06.012
Journal information: Structure
Provided by Chinese Academy of Sciences