This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Smart guide RNAs: Researchers use logic gate-based decision-making to construct circuits that control genes
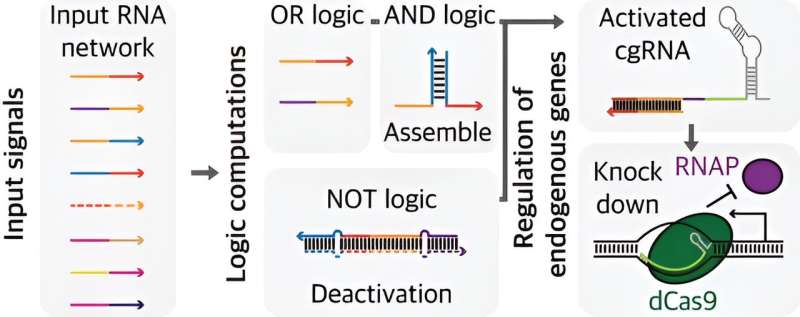
Researchers have transformed guide RNAs, which direct enzymes, into a smart RNA capable of controlling networks in response to various signals. A research team consisting of Professor Jongmin Kim and Ph.D. candidates Hansol Kang and Dongwon Park from the Department of Life Sciences at POSTECH has developed a multi-signal processing guide RNA.
This guide RNA can be programmed to logically regulate gene expression. Their findings were published in Nucleic Acids Research.
The CRISPR/Cas system, often referred to as "gene scissors," is a technology capable of editing gene sequences to add or delete biological functions. Central to this technology, which is used in several fields such as treating genetic diseases and genetically engineering crops, is a guide RNA that directs the enzyme to edit the gene sequence at a specific location.
While advances in RNA engineering have spurred research into guide RNAs that respond to biological signals, achieving precise control of networks of genes to respond to multiple signals has remained challenging.
In this study, the team combined the CRISPR/Cas system with biocomputing to overcome these limitations. Biocomputing is a technology that connects biological components like electronic circuits to program cellular and organismal activities.
The researchers implemented a guide RNA gene circuit capable of decision-making based on inputs, similar to a Boolean logic gate, which is one of the fundamental representations of input-output relationships in digitized signal operations.
The team successfully controlled essential genes involved in E. coli metabolism and cell division, demonstrating the capability to combine multiple logic gates for processing various signals and complex inputs. They used this circuit to control cell morphology and metabolic flows at the appropriate level.
This study is significant because it integrates existing systems and technologies to precisely control gene networks, enabling the processing, integration, and response to diverse signals within an organism. This goes beyond the role of guide RNAs in merely directing enzymes to specific locations.
Professor Jongmin Kim of POSTECH stated, "The research could enable the precise design of gene therapies based on biological signals within complex genetic circuits involved in disease. RNA molecular engineering allows for the simplicity of software-based structure design which will significantly advance the development of personalized treatments for cancer, genetic disorders, metabolic diseases, and more."
More information: Hansol Kang et al, Logical regulation of endogenous gene expression using programmable, multi-input processing CRISPR guide RNAs, Nucleic Acids Research (2024). DOI: 10.1093/nar/gkae549
Journal information: Nucleic Acids Research
Provided by Pohang University of Science and Technology