This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Functional predictability of universal gene circuits in diverse microbial hosts
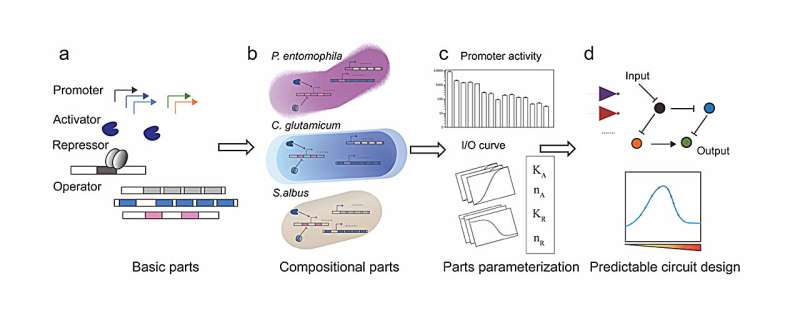
Over the past 20 years, synthetic biologists have been trying to build biological circuits in living cells to enact specific behaviors such as Boolean logic gates, signal filters, oscillators, state machines, sensors, and genetic controllers. The circuits have been built bottom-up from scratch by connecting genetic "parts" with certain commutating logics.
However, these achievements have largely been confined to a few model organisms like Escherichia coli and Saccharomyces cerevisiae. Transferring these genetic parts to non-model organisms is challenging due to host-specific gene expression mechanisms, metabolism, and varying DNA vectors.
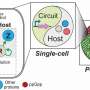
Thus, developing universal genetic circuits that can operate robustly and predictably across multiple organisms is crucial. The ideal universal genetic circuit should be insulated from the host environment, including extracellular, cellular, and genetic contexts.
A collaborative team from the Ouyang and Qian lab at Peking University and the Lou lab at Chinese Academy of Sciences published an article titled "Functional predictability of universal gene circuits in diverse microbial hosts" in the journal Quantitative Biology.
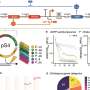
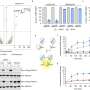
By developing a quantitative framework to explore the universality and reliability of biological parts in non-model organisms, the team characterized universal genetic parts, namely, the T7 RNA polymerase activation module and a set of transcriptional repression modules, in four microbial hosts including Streptomyces, Corynebacterium glutamicum, Pseudomonas putida, and Escherichia coli. Based on the excellent universality of these parts, the functional predictability of the genetic circuits was rigorously demonstrated.
The bottom-up design pipeline of genetic circuits across diverse species starts with the standardization of transcriptional regulatory elements originating from bacteria and bacteriophages, benchmarking these elements with single reaction models to ensure they perform consistently across host organisms.
Following this, parts are characterized combinatorically and parameterized through detailed modeling. Key parameters defining the behavior of these parts are extracted from measurements and categorized as intrinsic or host-specific. Finally, standardized parts are combined into complex circuits and combinatorial modeling and simulations are used to predict circuit behavior in different host organisms.
"These findings pave the way to predictably design and tune a universal genetic circuit with complex functions, and hold vast potential for realizing design automation of universal circuits in diverse hosts," writes Prof. Baojun Wang from Zhejiang University in a commentary published in the same issue.
More information: Chenrui Qin et al, Functional predictability of universal gene circuits in diverse microbial hosts, Quantitative Biology (2024). DOI: 10.1002/qub2.41
Provided by Frontiers Journals