This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers uncover details of how bacteria build protective barriers, may inform new antibiotics
Yale researchers have uncovered new details on how bacteria like E. coli build their protective barriers, which will inform new antibiotic development.
Antibiotic-resistant bacteria are a growing problem when it comes to combating infections. Bacteria that have an additional protective layer to their cell walls—a type known as "Gram-negative" in reference to the staining method used to identify it—are especially difficult to fight.
Yale researchers have made headway in understanding how bacteria generate this protective layer through a new study that uncovers additional nuance—and additional targets for developing new antibiotics.
Their findings were published April 18 in the Proceedings of the National Academy of Sciences.
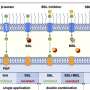
A critical component of this protective layer is a molecule called lipopolysaccharide (LPS). Bacteria need a certain amount of LPS; too much or too little kills the cell. Previous research from the lab of Wei Mi, assistant professor of pharmacology at Yale School of Medicine, revealed how molecular sensors in E. coli strike the right balance of LPS production.
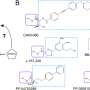
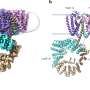
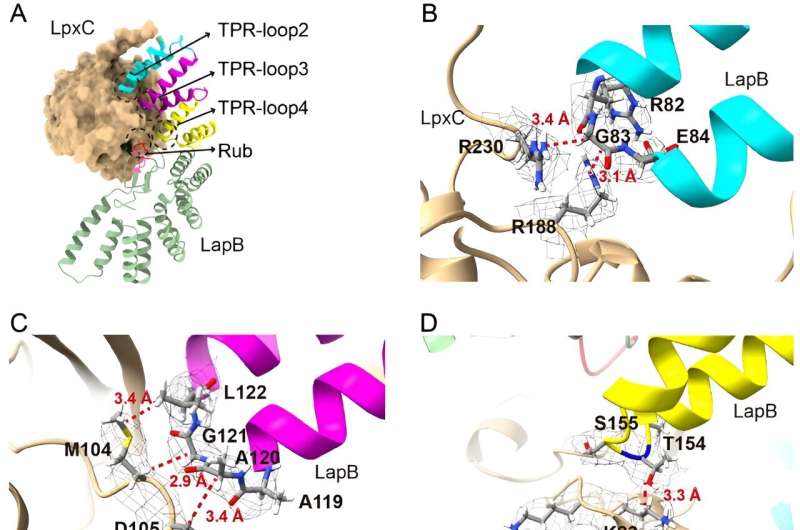
In the new study, researchers delved deeper into the part of this process that prevents excess LPS buildup, aiming to clarify how a protein called LapB binds to and stimulates the degradation of LpxC, an enzyme that kicks off LPS production. The researchers used cryogenic electron microscopy to view the structure of the complex created when these two molecules bind.
"Looking at the structure provides the most direct visualization of how this part of the process happens," said Mi, senior author of the new study. "Once we saw the structure, we made changes to the molecules to see how that affected binding, which allowed us to identify what components are necessary for LapB to recognize LpxC."
But the researchers also found, to their surprise, that LapB had a second role. Not only is it responsible for the degradation of LpxC, but it also inhibits the enzyme's action before degradation happens.
"Basically, LapB shuts down LpxC before it trashes it," said Mi. "We don't understand why bacteria do this, as it seems redundant, but this is what we're looking into now."
The researchers speculate that this dual role might be about flexibility. Degradation is a slow but irreversible process, whereas inhibition is prompt and reversible. Having both capabilities might enable bacteria to respond to environmental changes more nimbly.
"This is all relevant for antibiotic development," said Mi. "These details will help us find new approaches and understand why others don't work."
More information: Sheng Shu et al, Dual function of LapB (YciM) in regulating Escherichia coli lipopolysaccharide synthesis, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2321510121
Journal information: Proceedings of the National Academy of Sciences
Provided by Yale University