This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists generate new targeted protein degradation system that tunes a cell's own proteins
Researchers studying the role of proteins in health and disease use experimental tools that inactivate proteins, destroy them, or prevent them from being made in cells. In one approach, they mark targeted proteins with "destroy me" tags that work with small molecules known as molecular glues to prompt the cell's own protein-clearing machinery to gobble up the proteins. Yet, many tags used today are too large to tag the genes that encode a cell's native proteins, or they cause collateral damage, sparking destruction of proteins beyond the targeted one.
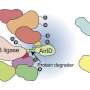
Now, scientists at the Broad institute of MIT and Harvard have used a continuous evolution platform called PACE to generate smaller protein degradation tags, or degrons, that form molecular glue complexes capable of precisely triggering depletion of a cell's own proteins.
The team then used a gene-editing technology they previously developed called prime editing to insert the compact degron into the genome of human cells, where it recruited cereblon, a key component of the cell's protein destruction pathway, in the presence of an otherwise-inert small molecule to rapidly induce the protein's degradation.
The scientists solved the three-component complex's 3D structure and revealed insights into its activity and specificity, and also evolved degron variants that work in mouse cells. The platform has the potential to evolve additional new degrons and molecular glue complexes that could be used to explore protein function or to validate therapeutic targets. The study appears in Science.
"We've shown that continuous evolution is a powerful approach to rapidly evolve molecular glue systems that can become useful research tools," said David Liu, senior author of the study and Richard Merkin Professor and director of the Merkin Institute of Transformative Technologies in Healthcare at the Broad.
"We're excited to see how the approach can also be applied to evolve other proteins that interact with small molecules in creative and powerful ways." Liu is also a Howard Hughes Medical Institute investigator and a professor at Harvard University.
New glue breakthrough
The effort builds on work published earlier this year and led by co-senior author Amit Choudhary, an associate member of the Broad and an assistant professor of medicine at Harvard Medical School.
That study identified a number of molecules related to thalidomide, a classic molecular glue that interacts with cereblon to trigger protein degradation and is used to treat some cancers and skin conditions. Whereas thalidomide triggers the destruction of numerous proteins, a related molecule known as PT-179 was relatively inert, making it a good starting point for a more precise molecular glue system.
In the new study, the researchers used a system called phage-assisted continuous evolution (PACE), first developed in Liu's lab at Harvard in 2011, to generate a degron that might work with PT-179. PACE allows researchers to rapidly evolve new proteins that have useful features.
In experiments led by co-first authors Jaron Mercer and Stephan DeCarlo, the team started with a zinc finger motif, a protein structural element known to engage molecular glue-bound cereblon. Through hundreds of generations of evolution during PACE, they evolved new zinc finger domains that interact with cereblon when bound to the PT-179 molecular glue.
The researchers then trimmed the zinc finger domain down to just 36 amino acids, dubbed SD40.
"We wanted to find the smallest motif possible, so that we could use our prime editing technology to efficiently tag endogenous genes in the genome," said DeCarlo, a graduate student at Harvard University and researcher in the Liu lab.
"It's a big goal in biology to study loss of proteins under endogenous regulatory control, not only for validating drug targets, but also for studying native protein function in biology."
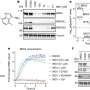
They then used prime editing to insert SD40 into the genome of human cells and tag two different proteins. The targeted proteins began disappearing from the cell mere minutes after adding PT-179, with no observed effect on any of thousands of non-targeted proteins evaluated.
"Other systems have shown degradation of a protein that's foreign to the cell, but using this potent tag that we evolved, we showed degradation of native target proteins from that cell, which is more biologically meaningful," said Mercer, a postdoctoral fellow in the Liu lab.
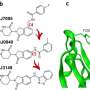
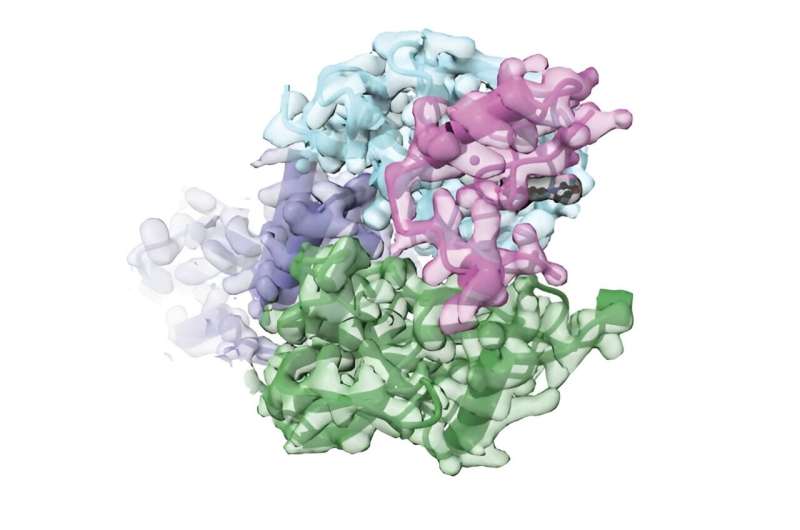
For a deep look at how SD40, cereblon, and PT-179 interact, the team analyzed the three-dimensional structure of the complex through work led by co-first author Shourya Roy Burman, a research fellow in the lab of co-senior author and cereblon expert Eric Fischer at Dana-Farber Cancer Institute.
Using cryo-electron microscopy, the team showed that when bound to SD40 and PT-179, cereblon is in a closed conformation, with SD40 appearing to hold the receptor's two ends together. The work suggests that cereblon may need to be "closed" in order to relay the message to other proteins in the complex to flag the targeted protein for destruction, although more work is needed to verify and understand this mechanism.
Since the cellular machinery that degrades proteins in human cells and in mice differs, the team also used their approach to evolve a degron that binds to cereblon found in mouse cells, which could be useful in studies involving transgenic mouse models of human disease.
The platform could also be used to evolve molecular glue complexes with novel degrons, or other combinations of proteins that interact in new, interesting ways.
The researchers are already working with scientists in cancer biology and other fields to harness their evolved degrons to tag proteins of potential therapeutic interest. This approach allows them to remove a target protein in minutes by adding PT-179 while preserving the corresponding gene's natural genomic context, thereby minimizing perturbation of regulatory mechanisms crucial to the native biological function of many proteins.
More information: Jaron A. M. Mercer et al, Continuous evolution of compact protein degradation tags regulated by selective molecular glues, Science (2024). DOI: 10.1126/science.adk4422
Journal information: Science
Provided by Broad Institute of MIT and Harvard