This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Scientists develop deep learning method to design bilin-binding proteins
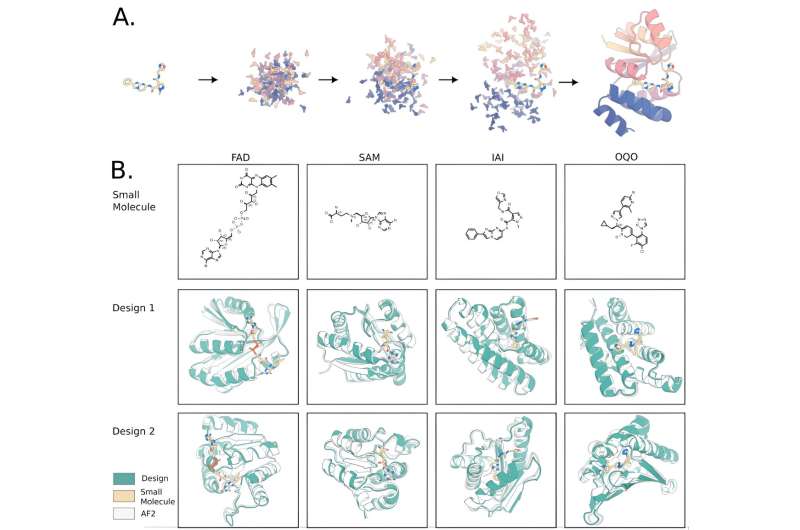
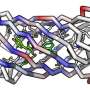
David Baker's group at the University of Washington, Seattle, U.S., have developed a novel deep learning method, RoseTTAFold All-Atom (RFAA), for prediction and design of complexes of proteins, small molecules, and nucleic acids. Subsequently, they developed RFdiffusionAA, which builds protein structures around small molecules.
These advances mean that in principle it is now possible to not only design a protein from scratch, but also to design proteins that will bind a range of cofactors and substrates. This advance would represent a breakthrough in protein design, because the majority of scientists work on proteins that bind small molecules of various types, but Baker's group needed a way to evaluate RFdiffusionAA.
Professor Neil Hunter, at the University of Sheffield, suggested bilins as a candidate small molecule, because his earlier work with former Ph.D. student Sam Barnett had furnished his group with E coli strains that synthesize bilins, and also make a native bilin-binding protein, CpcA. The study is published in the journal Science.
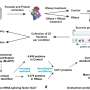
It was already known that bilins are optically featureless unless they are held within a defined binding site, at which point they become intensely colored and emissive. In this work current Ph.D. student in the Hunter/ Hitchcock group Felix Morey-Burrows devised a multiwell assay that could screen many RFdiffusionAA-generated genes in parallel, using E. coli cells that could make phycoerythrobilin (PEB).
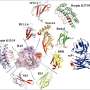
Morey-Burrows evaluated 94 designs in one go, with the multiwell assay revealing visibly colored cells in nine wells. He had identified nine proteins dissimilar to each other and to any native bilin binder, based on pigmentation or fluorescence. Most importantly, this assay proved that RFdiffusionAA works, and this method should be immediately useful for modeling protein-small molecule complexes, in particular multicomponent biomolecular assemblies for which there are few or no alternative methods available, and for designing small molecule binding proteins and sensors.
With regard to current work in the Photosynthesis Group at Sheffield, the 34/30 nm range in absorption/emission covered by just one design round using a single chromophore raises the exciting prospect of tailoring the spectral profiles of designed biliproteins by manipulating the conformational flexibility of the bilin and the protein microenvironment.
This work is not restricted to bilins, either. De novo designed antenna complexes could harvest light over a wider range of the UV-visible spectrum to enhance photosynthetic energy capture and conversion, and fluorescent reporter probes with tunable excitation/emission maxima would be useful biochemical tools.
More information: Rohith Krishna et al, Generalized biomolecular modeling and design with RoseTTAFold All-Atom, Science (2024). DOI: 10.1126/science.adl2528
Provided by University of Sheffield