This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Bacterial RNAs have shorter lifetimes than expected
The decay of ribonucleic acid (RNA) is a crucial mechanism for controlling gene expression in response to environmental stresses. Researchers from the Helmholtz Institute for RNA-based Infection Research (HIRI) and the Julius-Maximilians-Universität Würzburg (JMU) have developed a statistical approach that allows a more accurate prediction of RNA half-lives in bacteria.
Using this new method, the team discovered that the RNA half-life in Salmonella is three times shorter than previously assumed. Additionally, they shed further light on the role of RNA-binding proteins in RNA decay. The results were published in the Proceedings of the National Academy of Sciences.
Bacteria require specific proteins to multiply, and the blueprint for these proteins is embedded in their genetic material. Protein biosynthesis occurs in two steps: The information contained in DNA is transcribed into messenger ribonucleic acid (mRNA), which is then translated into proteins. In order to survive, microorganisms must rapidly adapt their proteome, the entirety of all proteins, to changing environmental conditions. Alongside transcription and translation, RNA decay is one of the key processes that control protein production.
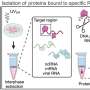
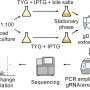
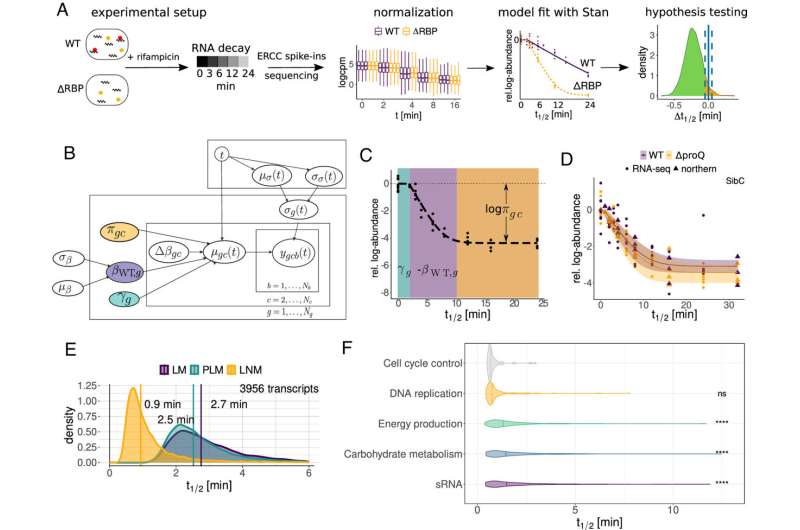
Common methods for determining the stability of bacterial RNA, such as the use of the antibiotic rifampicin, are prone to errors that can distort the derived results. Researchers from the Helmholtz Institute for RNA-based Infection Research (HIRI) and the Julius-Maximilians-Universität Würzburg (JMU) have developed a new approach to correct for these confounding effects.
"We combine high-throughput genomics data with a Bayesian statistical model. This Bayesian approach allows us to specify different hypotheses for how the data sets were generated. We can then directly test how well these competing hypotheses explain the observed data," says Lars Barquist. The computational biologist, who initiated the study, leads a research group at the Helmholtz Institute Würzburg, a site of the Braunschweig Helmholtz Center for Infection Research (HZI) in cooperation with the JMU. Barquist is also a professor at the University of Toronto in Canada.
"This method enables the analysis of RNA stability under different conditions or in mutant strains. It is a new and powerful tool for extracting biological parameters from complex data sets," elaborates Barquist.
Less time than anticipated
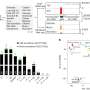
Using the new approach, the scientists investigated the half-life of RNAs in Salmonella enterica serovar Typhimurium. Surprisingly, they found that the half-life of the molecules is about three times shorter than previously thought—just under one minute instead of approximately three minutes. The data from the study suggest that the half-life of bacterial RNAs in general have been significantly overestimated. This is likely to lead to a broader reassessment of decay rates for other bacteria, as they have typically been determined using the rifampicin RNA stability assay.
"RNA decay rates are one of the fundamental parameters governing gene expression. Our findings have implications for almost all studies dealing with the adaptation of the transcriptome, the total amount of all transcribed genes, and the proteome of bacteria to different environments, such as during infection or after antibiotic treatment," explains Laura Jenniches, postdoctoral researcher in Barquist's group and first author of the study.
A new perspective on proteins
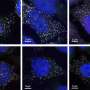
The new technique also gave the team the opportunity to look more closely at the interactions between RNA-binding proteins (RBPs) and RNA decay. While these proteins are known to be involved in post-transcriptional regulation in bacteria, their influence on RNA half-life has not been thoroughly studied.
The researchers identified significant transcript cohorts whose stability changed after RNA-binding proteins were knocked down. These observations provide new insights into the role of RBPs in shaping the transcriptome and suggest that these proteins play overlapping roles in maintaining cellular balance.
"Our method allows us to examine the activity of the RBP network in more detail and gain insights into how proteins control responses to stress. This study is a first step towards a better understanding of global regulation coordinated by RBPs in bacterial pathogens," says Barquist.
More information: Laura Jenniches et al, Improved RNA stability estimation through Bayesian modeling reveals most Salmonella transcripts have subminute half-lives, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2308814121
Journal information: Proceedings of the National Academy of Sciences
Provided by Helmholtz Association of German Research Centres