This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Creating a toolkit of yeast strains that over-produce key cellular building blocks
Microbes such as bacteria and yeast are increasingly being used to produce components of medicines, biofuels, and food. Indeed, baker's yeast, also known as brewer's yeast or Saccharomyces cerevisiae, is responsible for the fermentation process used in making beer or bread, but it is also used at scale to produce other molecules of value for industry.
Using microbes could be key in producing these materials more sustainably, but there is still a lot we don't understand about how microbial communities—particularly yeast—form and keep going.
To answer these questions, Imperial College London researchers created a molecular toolkit that uses a new way to produce useful compounds. Their work is published in the journal Nature Microbiology.
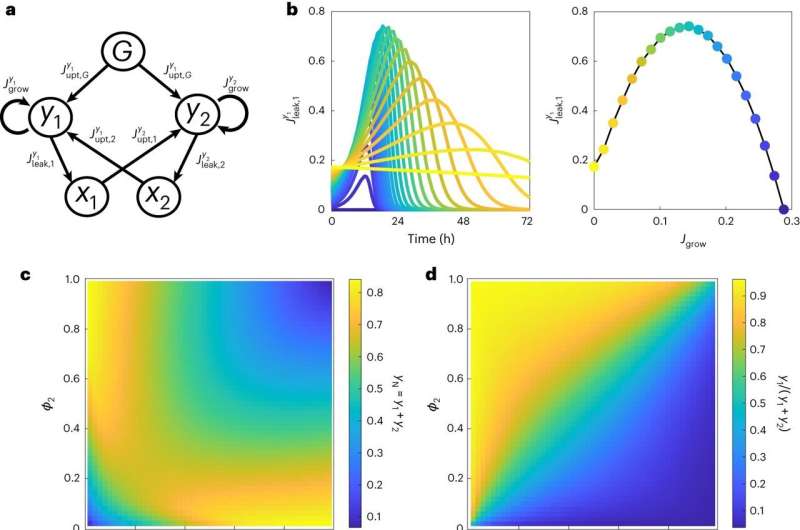
The toolkit consists of 15 different yeast strains that over-produce key cellular building blocks—amino acids and nucleotides—but lack the ability to make other building blocks. Unlike traditional synthetic biology communities, the strains were split into 'donors'—strains that donate growth building blocks to others, and 'receivers'—strains that receive them.
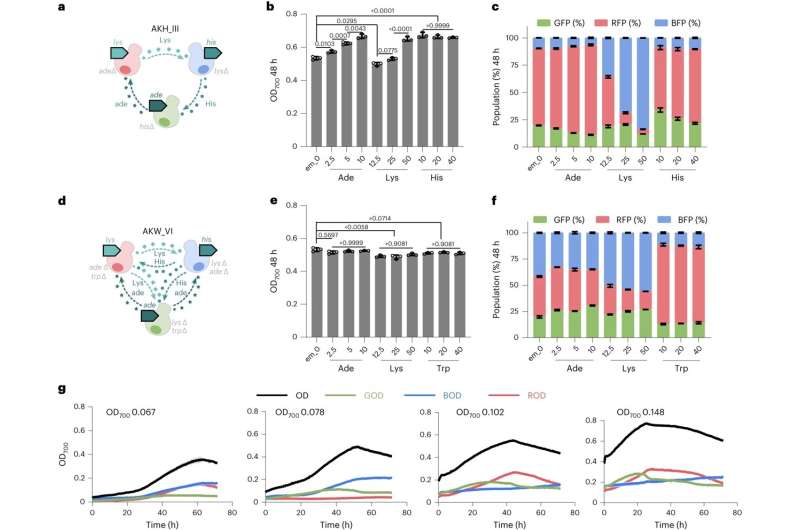
To test the kit, the researchers investigated the effectiveness of the donor-receiver system on communities' ability to produce resveratrol, an antioxidant compound found in red wine, and some food supplements that are being investigated for their potential medicinal properties.
Using the new donor-receiver system, the researchers created yeast communities with two or three different strains each and observed how they interacted and grew together. The toolkit allowed them to split the resveratrol production pathway in two, placing each half in a selection of donor and receiver yeast strains.
As a control, they also produced resveratrol using standard methods with just one yeast strain.
They then measured the effects of variables such as adding extra nutrients, altering the initial mix of different yeast strains, and altering how densely packed the cells were on the communities' behavior and the rate at which they produced resveratrol.
They found that spitting the pathway between two yeast strains resulted in enhanced production of resveratrol when compared to the traditional production platform. Mathematical modeling showed that the system also allowed for more stable and specific partnerships between the yeast strains.
Senior author Dr. Rodrigo Ledesma-Amaro, from Imperial's Department of Bioengineering, said, "Our findings, if replicated across further yeasts and metabolites, have the potential to significantly impact the way we understand and use microbial communities in sustainable bioproduction, from food to biofuels."
Although tested using one strain producing one compound so far, the new system could allow researchers to create various combinations of yeast communities that work together and make various products more efficiently and sustainably.
The ability to engineer and boost the efficiency of yeast communities could improve our production of pharmaceuticals, food, beverages, bioplastics, and biofuels. Efficiency could also lead to less waste, lower energy consumption, and lower costs in producing valuable compounds.
First author Dr. Huadong Peng, who conducted the work at Imperial's Department of Bioengineering, said, "Our study is the first of its kind to use both mathematical modeling and practical experiments to understand how factors inside and outside the yeast cells affect community growth. Our findings present exciting possibilities for further study."
Next, the researchers will refine the toolkit and expand its scope to include a wider range of small molecules beyond the current 15 amino acids and nucleotides. They will also lengthen the testing to understand the kit's long-term stability, which will be essential for practical applications where these communities might be used for prolonged periods. They will also introduce different types of yeast and monitor for mutations and adaptations that might affect results.
More information: Huadong Peng et al, A molecular toolkit of cross-feeding strains for engineering synthetic yeast communities, Nature Microbiology (2024). DOI: 10.1038/s41564-023-01596-4
Provided by Imperial College London




















