This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Supercomputing the secrets inside cattle antibiotics
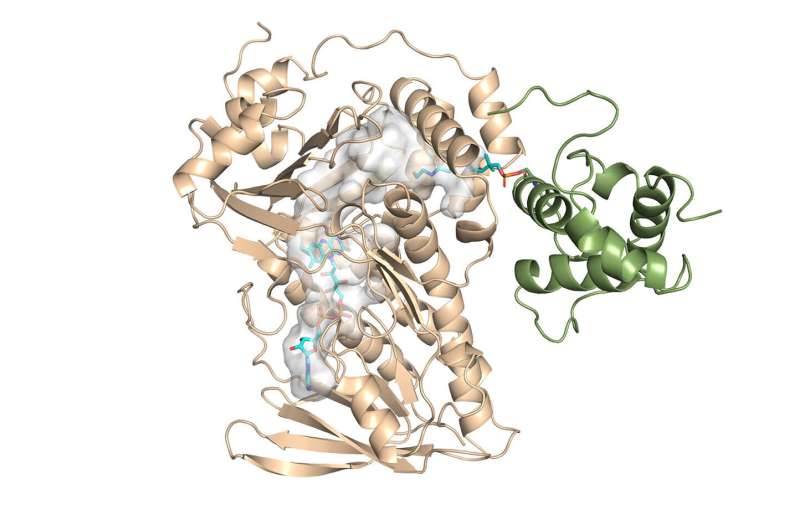
Chemists have determined for the first time the crystal structure of a key component of the monensin enzyme and have unlocked the mechanism of its reaction activity.
"The main finding was the first crystal structure for this family of enzymes," said Chu-Young Kim, a professor of Biochemistry at the University of Illinois Urbana-Champaign (UIUC), who led the experimental side of the study. He and colleagues solved the crystal structure for MonCI, a key enzyme in soil bacteria that naturally synthesizes monensin.
Lela Vukovic, an associate professor at The University of Texas, at El Paso (UTEP), performed the computational studies on the monensin research, published in Nature Communications.
The University of Texas Research Cyberinfrastructure (UTRC) initiative awarded Vukovic supercomputer allocations on the Lonestar6 system at the Texas Advanced Computing Center (TACC) to meet these challenges. UTRC provides advanced computing capabilities to researchers across all 14 UT System institutions.
TACC's Lonestar6 helped reveal the reaction sequence that produces monensin. This research opens the door for future design of safer, more effective antibiotics.
"We found that MonCI is used to carry out three crucial epoxidation reactions," Kim said. "This is very unusual and has implications on how we can engineer the bacterium to produce new antibiotics."
Kim, who recently left UTEP to join UIUC, consulted Vukovic's lab with the structure results and full of new questions. What he found was an interesting sequential reaction inside the enzyme. However, it was still experimentally impossible to obtain the crystal structure of the enzyme with the substrate inside in its active mode.
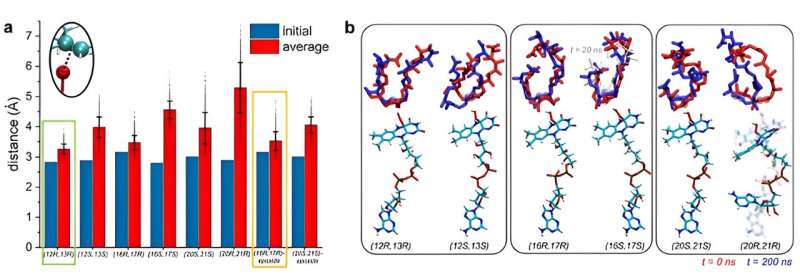
That's when Kim and Vukovic decided to model the enzyme and substate in simulations when the substrate is stable. If a substrate is stable in a certain position, then a reaction can occur for that position.
Vukovic and her students Tara Nitka and Anju Yadav developed full models of the roughly 78,000-atom system based on the crystal structures determined by the study experimentalists.
"The computational challenges arose from examining multiple systems to determine the position in which premonensin A and its epoxidated versions are most stable and most likely to undergo first, second, and third epoxidation reactions," Vukovic said.
"Supercomputers have been very useful in characterizing these biological molecules that naturally produce antibiotics," Vukovic added. "We would not be able to perform the computational studies without them. Computational studies help us discover and understand these complex sequential reactions important to society."
Vukovic performed her postdoctoral research at UIUC under the late Klaus Schulten, whose legacy lives on in the NAMD software his group developed and was used in this study and countless others.
"UIUC is doing a lot of work to optimize NAMD to run on the nation's supercomputers, such as Lonertar6 and Stampede2 at TACC," she said. "NAMD allowed us to zoom into this enzyme and see which reactions happen first, second, and third to generate monensin."
"Monensin biosynthesis requires at least 14 different enzymes, one of which is MonCI," Kim added. "We must also investigate all of the other enzymes. In the future, we expect to generate improved versions of monensin to better take care of cattle and poultry. Additionally, monensin is toxic to horses and dogs, so these farm animals are sometimes accidentally poisoned and killed. Therefore, a nontoxic monensin is needed."
The authors are Qian Wang, Tara A. Nitka, Anju Yadav, and Lela Vukovic of The University of Texas at El Paso; Ning Liu, Hongli Xiao, Hui Yang, and Xi Chen of Northwest University, China; Irimpan I. Mathews of the SLAC National Accelerator Laboratory; and Chu-Young Kim of the University of Illinois Urbana-Champaign.
More information: Qian Wang et al, Triepoxide formation by a flavin-dependent monooxygenase in monensin biosynthesis, Nature Communications (2023). DOI: 10.1038/s41467-023-41889-0
Provided by University of Texas at Austin