This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Q&A: Research visualizes a precise mechanism for how cells sort their trash
For decades it has been an open question in the ubiquitin research field how proteins are labeled as being defective or unneeded. In a recent study Brenda Schulman, Director at the Max Planck Institute (MPI) of Biochemistry, and Gary Kleiger, Chair of Chemistry and Biochemistry Department at University of Las Vegas Nevada, together with their teams were able to visualize this precise mechanism, catalyzed by the Cullin-RING Ligase E3s, for the first time.
In an interview, Brenda Schulman explained what led them to their findings, as well as how this knowledge may be used to help treat diseases. The results of their study are published in the journal Nature Structural & Molecular Biology.
Cullin-RING-ligases (CRLs) are complex nanomachines that are crucial for the cell's intricate disposal and recycling systems. CRLs tag defective, toxic, or superfluous proteins with repeating units of a small protein called ubiquitin, forming a chain of ubiquitins in a process called poly-ubiquitin labeling.
By doing so, CRLs mark the protein as "to be degraded" and prevent the toxic accumulation of unnecessary proteins. When this process fails, cellular trash can build up. So mutations or misfunctions impairing CRLs are often associated with diseases, like developmental disorders or cancers. Because of their key functions in maintaining the well-being of our cells, it is of fundamental importance to define and understand their molecular mechanisms.
Through a method called cryo Electron Microscopy, or cryo-EM for short, it is now possible to visualize some of the tiniest structures of life, such as CRLs, thus providing key insight into their functional states.
Brenda, you've dedicated half your life to research in the ubiquitin field. Can you tell us what exactly fascinated you so much about it?
Ubiquitin is a fascinating protein, especially considering its small size compared to most other proteins. Its name comes from 'ubiquitous' which means found everywhere. Indeed, we now know that ubiquitin is nearly everywhere—in plants, fungi, insects, animals and humans. And even though bacteria and viruses do not make ubiquitin, they hijack our ubiquitin to promote infections.
Ubiquitin's action is controlled by a system of hundreds of different molecular machines, called E3 ligases, which dictate where and when ubiquitin is attached to the cell's trash. Since ubiquitin is essentially ubiquitous, E3 ligases play important roles in turning on and off of most cellular processes.
Studying ubiquitin for over 25 years now is like being a master detective in a long-running mystery series, especially since E3 ligases do essentially pull the trigger for eliminating other proteins.
After we solve one mystery, there is always another with even bigger challenges. And the technologies in our forensics toolbox—for example, CRISPR, chemical biology and cryo-EM, to name a few—have become far more sophisticated. So it remains both a thrilling adventure and fulfilling to solve these mysteries together with my colleagues.
Your recent study discovered how a lot of different cellular proteins are marked with ubiquitin chains—very rapidly—when they need to be eliminated. What led you and your team to this discovery?
Let me mention that this work is really gratifying because this has been a mystery for decades, and it is one I set out to solve when I started my first independent group 23 years ago.
Our collaborator, Gary Kleiger, at the University of Nevada, Las Vegas showed that a CRL picks out the target for labeling, holds it in place while attaching poly-ubiquitin, and then releases the target so the CRL can then label another target for destruction, all within milliseconds. In fact, this breakneck efficiency explains why so far it has not been possible to take pictures of this process—it is too fast to capture using available techniques. We had to develop a whole new method to visualize how CRLs label proteins for destruction.
Can you tell us more about the method you've developed to reveal the mechanism?
We developed a chemical tool so that when the CRL starts to label a protein with poly-ubiquitin, the chemical acts like a trap, halting the CRL during the key moment when the poly-ubiquitin label is being attached to the protein. This allowed us to visualize the complex by a structural biology method called cryo-EM.
Seeing all the molecular components coming together led to discovering the mechanism of poly-ubiquitin labeling catalyzed by CRLs. Next, our longtime collaborator Gary Kleiger and his lab developed a new assay to validate the model, which monitored these reactions in milliseconds—the first time-point they detect is 1/400th of a second!
What were the most promising observations that you think will play a key role in the future development of the field?
Thanks to our chemical trap we obtained a variety of different snapshots of CRLs "in action." These informed how poly-ubiquitin labeling works on both naturally occurring proteins in cells, and on one when a drug-like molecule, MZ1, is applied. MZ1 triggers disposal of a cancer-associated protein. All of these showed us a lot of interesting facts, but there are three main findings.
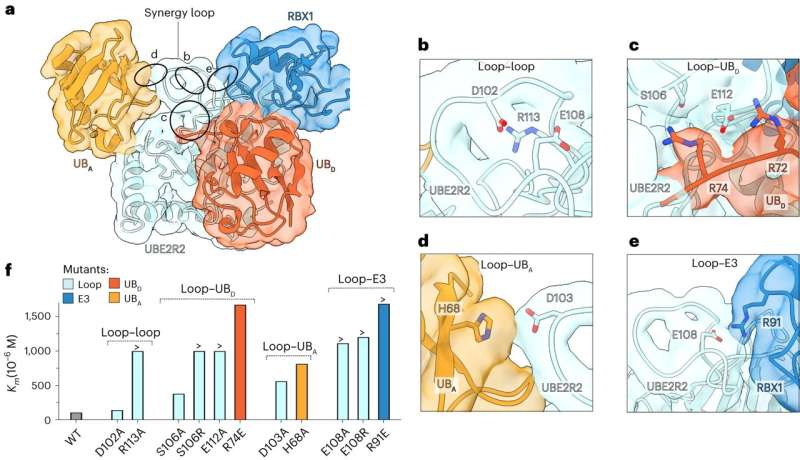
The first was the revelation of the actual reaction mechanism of poly-ubiquitin labeling. Our structural snapshots showed all molecular components synergizing for ultra-rapid and specific catalysis. Indeed, speed and specificity are the hallmark features of enzymes! This has been a question in the field for over 30 years now and we are thrilled to be the first ones to show the mechanism of this shockingly fast, critical reaction.
The second key observation came from comparing the new data to our previously published work on how CRLs execute another, earlier, step in the tagging of proteins for disposal. This earlier step involves initial placement of one ubiquitin, the new study shows how the poly-ubiquitin label that contains multiple ubiquitins is made. Crucial parts of the CRL molecular machine are drastically rearranged during the former and latter processes.
Perhaps most important was our finding that these features were observed for all the CRLs that we examined labeling proteins with poly-ubiquitin tags, suggesting that our mechanism may apply to the suite of CRLs in our cells.
What do you expect your findings to be used for in the future?
First of all, this is a big leap in the ubiquitin field that changes the way we think about a ubiquitous cellular system. We now know that all the parts of the poly-ubiquitin labeling machinery have to come together in a specific way for the reaction to work with blazing speed and exquisite specificity.
Second, our study has implications for the emerging drug development field called targeted protein degradation (TPD). Most TPD efforts use small molecules to recruit disease-causing proteins to CRLs. This leads to poly-ubiquitin labeling and ultimately the elimination of the disease-causing protein. The new data indicate molecular and geometric constraints for employing CRLs for TPD, which we showed for one of the pioneering degrader molecules, MZ1.
This field is very exciting and a complementary preprint on CRL targeting via MZ1 was also posted on bioRxiv by the lab of Alessio Ciulli, who developed MZ1. Our findings could help to develop new TPD drugs and understand how they work in diseased cells.
More information: Joanna Liwocha et al, Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases, Nature Structural & Molecular Biology (2024). DOI: 10.1038/s41594-023-01206-1
Charlotte Crowe et al, Mechanism of degrader-targeted protein ubiquitinability, bioRxiv (2024). DOI: 10.1101/2024.02.05.578957
Journal information: Nature Structural & Molecular Biology , bioRxiv
Provided by Max Planck Society