This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Newly identified protein helps flowers develop properly
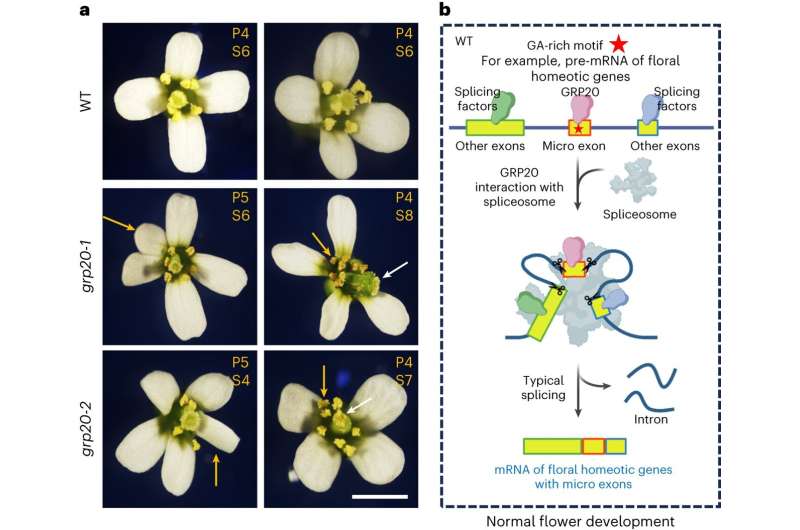
Flowers rely on a newly identified protein to develop properly with all of their organs, according to the research team who made the discovery. The team, led by Penn State biologists, identified the protein in the model plant species Arabidopsis and said its mechanism is likely shared across plant species.
They found that the protein helps ensure RNAs—molecules that transfer information stored in DNA to produce proteins—in flowers are properly processed. It does this by keeping even the smallest components of RNA code from becoming accidentally lost before they can be used to make corresponding proteins. Some of these proteins are, in turn, needed for the normal formation of all floral organs, including petals and stamens.
The researchers demonstrated that the protein, required for proper flower development, is involved in processing of RNAs for thousands of genes and probably helps the plant respond to the environment.
A paper describing the research appeared in the journal Nature Plants.
"The protein we identified in Arabidopsis, called GRP20, helps ensure that thousands of genes are properly spliced and even the smallest exons are not missed in this splicing process for crucial floral regulatory RNAs," said Hong Ma, holder of the Huck Chair in Plant Reproductive Development and Evolution and professor of biology in the Eberly College of Science at Penn State, who led the research team.
RNA can have their own function or carry genetic code used to generate proteins. However, for most genes, not all of the sequence contained in the RNA is used for coding proteins. These RNA molecules must first be processed, a step called splicing, cutting out noncoding bits and rejoining together the coding parts, called exons.
Splicing is carried out by the spliceosome—molecular machinery containing multiple proteins and RNA molecules—and is further regulated by other splicing-factor proteins. The average length of an exon in plants is 180 nucleotides—the A, T, C, G units that compose DNA.
These exons usually contain specific sequences of nucleotides that are recognized by the splicing machinery, but micro exons comprising 50 or fewer nucleotides often lack these sequences.
"Relatively little is known about the mechanisms that control the splicing process for specific sets of RNAs beyond the general process by the spliceosome, especially for those with micro exons," Ma said. "Short exons, because of their length, can be missing particular sequences that guide proper recognition of the exons by the splicing machinery."
In addition to the spliceosome and known splicing factors, researchers predicted additional specialized proteins exist to ensure that the short exons are not missed in the splicing process.
"GRP20 both binds to RNA and interacts with the spliceosome," Ma said. "We experimentally disrupted the gene encoding this protein in the plant and found that over 2,000 genes were improperly spliced during the plant's development. We specifically saw that micro exons were missing in genes known to be important for floral organ formation and response to environmental change."
The researchers identified which part of the GRP20 protein binds to the RNA molecules and which part interacts with the spliceosome. They then created versions of the GRP20 protein that lacked either the RNA binding capacity or the ability to interact with the spliceosome.
"In both cases, the plants failed to develop properly due to missing micro exons in RNAs coding for proteins important in flower development," Ma said. "This suggests that both RNA binding and interaction with the spliceosome are required for the proper function of the protein in ensuring the proper inclusion of the micro exons in RNAs for normal flower formation."
The researchers further showed that they could rescue flower development by introducing proteins that contained all of the portions coded by micro exons, or by introducing a version of the GRP20 gene from another plant species that is a close relative of cabbage and canola.
"The discovery that GRP20 is a crucial regulator for RNA splicing will allow us to further explore its function in the regulation of other molecular, developmental and physiological plant processes," Ma said. "Additionally, because the GRP20 protein and many of the genes that it helps to properly splice are conserved across plant species, we suspect that its role is also conserved and plan to explore its function, and possibly that of other similar proteins, in other species."
In addition to Ma, the research team includes Jun Wang, Xinwei Ma, Yi Hu, Guanhua Feng and Xin Zhang at Penn State; and Chunce Guo at Jiangxi Agricultural University in China. The Penn State Eberly College of Science and the Penn State Huck Institutes of the Life Sciences supported this research.
More information: Jun Wang et al, Regulation of micro- and small-exon retention and other splicing processes by GRP20 for flower development, Nature Plants (2024). DOI: 10.1038/s41477-023-01605-8
GRP20 regulates micro-exon retention via interaction with the spliceosome during flower development, Nature Plants (2024). DOI: 10.1038/s41477-023-01606-7
Journal information: Nature Plants
Provided by Pennsylvania State University