This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists reveal how RNA gets spliced correctly
To carry out all of life's functions, proteins must be produced from instructions carried by genes within DNA and delivered to the cell's protein-making machinery by messenger RNA.
However, to generate mature mRNA, intervening sequences called introns must be removed through a process called splicing. Errors that occur during splicing can potentially cause disease.
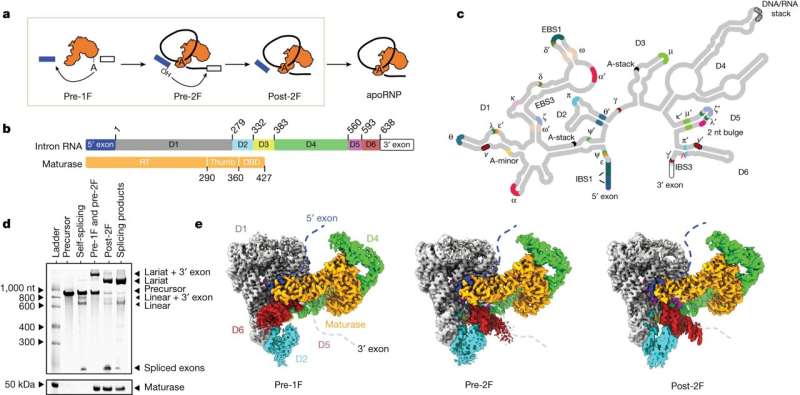
In a new study published in the journal Nature, a research group headed by the lab of Anna Marie Pyle, Sterling Professor in the Departments of Molecular Cellular and Developmental Biology and Chemistry at Yale and Investigator of the Howard Hughes Medical Institute, explored mechanics of the splicing process. To do so, they studied an ancient ancestor of the spliceosome, a large complex of proteins and RNA that cuts out intervening sequences.
"Every gene contains introns that must be removed in a conserved process carried out by the spliceosome," said Ling Xu, a postdoctoral fellow in the Pyle lab and lead author of the study. "And we found that these mechanisms are shared by organisms from bacteria to humans."
Writing in Nature, the authors describe the intricate series of biochemical and structural changes that enable intron removal.
"These are highly regulated actions and the key components, and the fundamental chemistry of splicing haven't changed from ancient times to now," said Tianshuo Liu, a graduate student in Yale's Department of Molecular, Cellular, and Developmental Biology and co-author of the study.
"And whenever a mistake occurs during splicing, you will find a disease as a result," added Kevin Chung, a graduate student in the Pyle lab and co-author.
Aberrant splicing of mRNA has been implicated in neurodegenerative and neuromuscular diseases such as Parkinson's and spinal muscular atrophy.
More information: Ling Xu et al, Structural insights into intron catalysis and dynamics during splicing, Nature (2023). DOI: 10.1038/s41586-023-06746-6
Journal information: Nature
Provided by Yale University