This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Study finds a smoking gun for the spread and evolution of antibiotic resistance
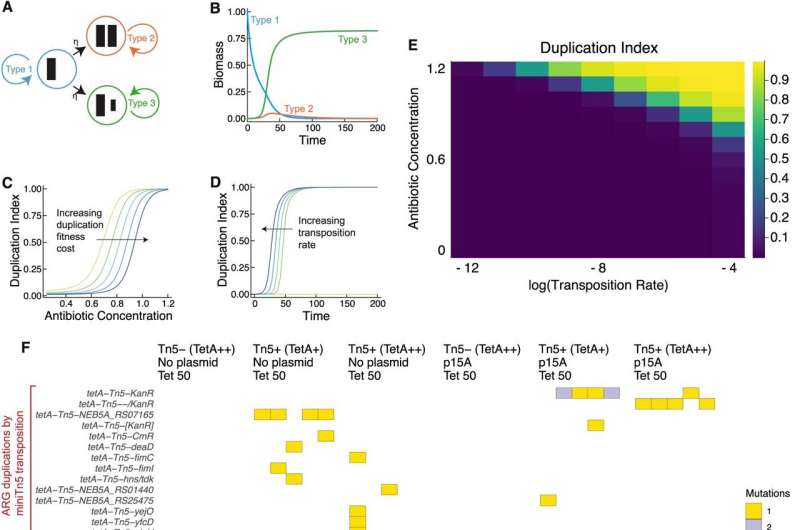
Biomedical engineers at Duke University have uncovered a key link between the spread of antibiotic resistance genes and the evolution of resistance to new drugs in certain pathogens.
The research shows bacteria exposed to higher levels of antibiotics often harbor multiple identical copies of protective antibiotic resistance genes. These duplicated resistance genes are often linked to "jumping genes" called transposons that can move from strain to strain. Not only does this provide a mechanism for resistance to spread, having multiple copies of a resistance gene can also provide a handle for evolution to generate resistance to new types of drugs.
The results appear in the journal Nature Communications.
Earlier work by the Lingchong You lab has shown that 25% of bacterial pathogens are capable of spreading antibiotic resistance through horizontal gene transfer. They have also shown that the presence of antibiotics does not speed up the rate of horizontal gene transfer, so there must be something else happening that pushes the genes to spread.
"Bacteria are constantly evolving under many pressures, and elevated duplication of certain genes is like a fingerprint left at the crime scene that allows us to see what kinds of functions are evolving really rapidly," said Rohan Maddamsetti, a postdoctoral fellow working in the laboratory of Lingchong You, the James L. Meriam Distinguished Professor of Biomedical Engineering at Duke. "We hypothesized that bacteria under attack from antibiotics would often have multiple copies of protective resistance genes, but until recently we didn't have the technology to find the smoking gun."
Traditional DNA-reading technology copies short snippets of genes and counts them up, making it hard to determine whether high counts of specific sequences are actually in the sample or if they are being artificially amplified by the reading process. In the past five years, however, complete genome sequencing with long-read technology has become more common, allowing researchers to spot high levels of genetic repetition.
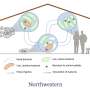
In the study, Maddamsetti and co-authors counted the repetitions of resistance genes present in samples of bacterial pathogens taken from a variety of environments. They discovered that those living in places with higher levels of antibiotic use—humans and livestock—are enriched with multiple identical copies of antibiotic resistance genes, while such duplications are rare in bacteria living in wild plants, animals, soil and water.
"Most bacteria have some basic antibiotic resistance genes in them, but we rarely saw them being duplicated out in nature," You said. "By contrast, we saw lots of duplication happening in humans and livestock where we're likely hammering them with antibiotics."
The researchers also found that the levels of resistance duplication were even higher in samples taken from clinical datasets where patients are likely taking antibiotics. This is an important point, they say, because the increase in copying antibiotic resistance genes also increases the likelihood of bacteria evolving resistance to new types of treatments.
"Constantly creating copies of genes for resistance to penicillin, for example, may be the first step toward being able to break down a new kind of drug," Maddamsetti said. "It gives evolution more rolls of the dice to find a special mutation."
"Everyone recognizes there is a growing antibiotic resistance crisis, and the knee jerk reaction is to develop new antibiotics," You added. "But what we find time and again is that if we can figure out how to use antibiotics more efficiently and effectively, we can potentially address this crisis much more effectively than simply developing new drugs."
"The majority of antibiotics used in the United States are not used on patients, they're used in agriculture," You added. "So this is an especially important message for the livestock industry, which is a major driver of why antibiotic resistance is always out there and becoming more serious."
More information: Rohan Maddamsetti et al, Duplicated antibiotic resistance genes reveal ongoing selection and horizontal gene transfer in bacteria, Nature Communications (2024). DOI: 10.1038/s41467-024-45638-9
Provided by Duke University