This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Mouse study finds genetic variation determines the actions of gene regulatory factors
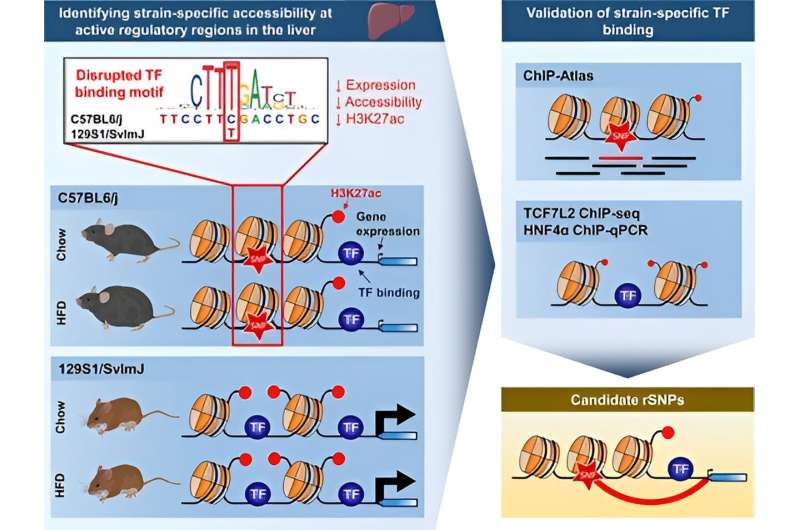
A recent study from the University of Eastern Finland shows that genetic variation determines chromatin accessibility and the binding of transcription factors. Chromatin consists of DNA and proteins attached to it. The density of this packaging is regulated by transcription factors, and DNA accessibility is required for gene expression. The research is published in the journal Nucleic Acids Research.
Most of the genetic variation that increases the risk of complex diseases targets regions outside the protein-coding areas of genes. Transcription factors require binding to a specific DNA sequence within chromatin, and changes to this sequence can prevent binding, thereby influencing gene expression. Chromatin accessibility is often used to identify binding sites for regulatory factors.
The study compared the activity of regulatory regions of genes and gene expression in the liver, as well as responses to a high-fat diet in two genetically distinct mouse strains. These mouse strains are commonly used in studying obesity susceptibility and type 2 diabetes.
No differences were observed within the mouse strains when comparing chromatin accessibility with a regular and high-fat diet. However, significant differences in chromatin accessibility were found between the mouse strains, particularly at chromosomal locations where genetic differences between the strains were located. Moreover, these differences often occurred near genes that were expressed differently between the strains. Thus, genetic variation affected the binding of transcription factors, and these changes were reflected in gene expression.
The study also examined how reliably changes caused by genetic variation in the binding sites of individual transcription factors could be predicted and what type of genomic information this would require.
The combination of data describing chromatin accessibility, genetic variation and binding regions of transcription factors provided the best prediction of the binding of transcription factors directly affecting chromatin accessibility. However, the reliability of the prediction was compromised in those cases where the same genetic variation affected the binding sequences of several transcription factors. Therefore, the impact of genetic variation on each transcription factor's binding must be measured directly.
"This research deepens our understanding of the connections between genetics, modifications to chromatin, and transcription factor actions that direct gene expression. Additionally, the study describes how public datasets can be utilized in investigating the mechanisms behind the hereditary risk factors for complex diseases," says University Teacher Juho Mononen of the University of Eastern Finland.
More information: Juho Mononen et al, Genetic variation is a key determinant of chromatin accessibility and drives differences in the regulatory landscape of C57BL/6J and 129S1/SvImJ mice, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad1225
Journal information: Nucleic Acids Research
Provided by University of Eastern Finland