This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Enhancing cultivated strawberry diversity and disease resistance
Cultivated strawberries are now the basis of a multi-billion-dollar global industry. To meet the challenges posed by increasing demand, climate change, and water and land scarcity, new germplasm needs to be collected to improve cultivars. However, managing clonally propagated germplasm collections such as strawberry is costly, while limited genotypic and phenotypic characterization hinders effective germplasm utilization.
Genome assembly and genotyping platforms have enabled a deeper understanding of genetic diversity and trait genetics. The USDA-ARS National Clonal Germplasm Repository (NCGR) in Corvallis, Oregon maintains the U.S. strawberry collection, yet very little has been done to explore the molecular diversity within the U.S. National F. ×ananassa collection .
In May 2022, Horticulture Research published a research article titled "Exploring the diversity and genetic structure of the U.S. National Cultivated Strawberry Collection ."
In this study, data curation involved genotyping strawberry accessions using the Axiom IStraw35 and Axiom FanaSNP arrays, which identified 27,968 and 40,424 SNP markers, respectively. The marker data shared by the two arrays were analyzed for call quality, missing data, and minor allele frequencies, resulting in 4,033 markers for structural assessment, diversity analysis, pedigree confirmation, core collection development, and identification of haplotypes associated with desirable traits.
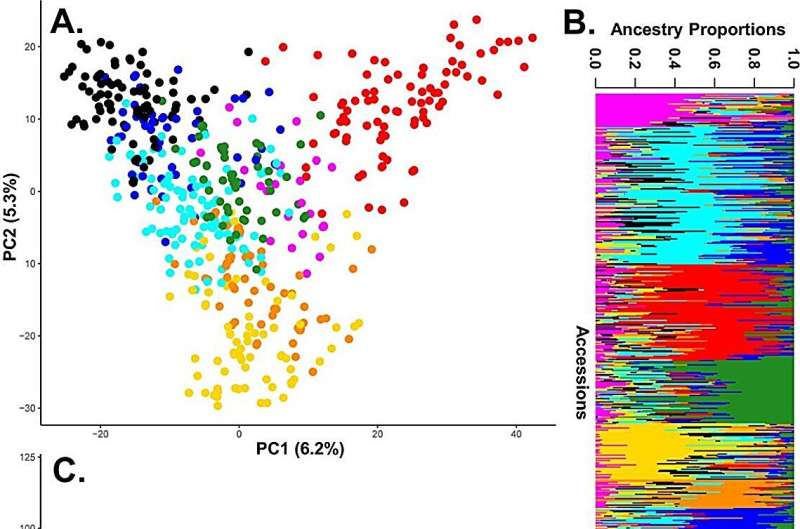
Population structure was analyzed using Principal Component Analysis (PCA). K-means clustering and the sNMF algorithm both identified eight optimal subpopulations. F. ×ananassa had the same diversity across geographic regions. Hierarchical clustering using UPGMA identified seven to nine major clades that were also correlated with geographical strata.
STRUCTURE analysis identified three broad subgroups with diversity statistics showing similar richness and evenness across geographic populations. Two core collections were established, type 1 core collection (CC-I) and type 2 core collection (CC-X) , each comprising 100 individuals. These collections were significantly diverse compared to random sampling, represented major geographic regions and all groups from the population structure analysis.
The COLONY output confirmed pedigree links for 241 of 308 accessions, with 78.0% appearing true-to-type. Trait-associated haplotype prevalence analyses showed that only 3,943 of the 4,033 markers mapped uniquely to the F. ×ananassa "Camarosa" assembly. Marker density was insufficient in certain regions to identify trait-associated haplotypes. Notably, some accessions like "Fletcher" and "L'Amour" possessed multiple resistance haplotypes. Finally, accessions containing FaRCa1, FaRCg1, FaRMp1, and FaRPc2 resistance-related haplotypes were identified.
In summary, germplasm collections provide a wealth of resources for breeders to develop and improve cultivars. The core collections created in this study will be useful tools for identifying new genes and validating DNA information for breeding. These findings are valuable for breeding and research, providing a more efficient way to utilize the F. ×ananassa collection.
More information: Jason D Zurn et al, Exploring the diversity and genetic structure of the U.S. National Cultivated Strawberry Collection, Horticulture Research (2022). DOI: 10.1093/hr/uhac125
Journal information: Horticulture Research
Provided by Plant Phenomics