December 5, 2023 dialog
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
written by researcher(s)
proofread
Circadian stress response provides an insight into metabolic communication via the mitochondrial epigenome
Who needs science fiction when you have the mitochondria? Billions of years ago, early plant and animal cells were infected by protobacteria which sought refuge from the outside world. Over time, these bacteria formed a symbiosis with our ancestors, removing damaging oxygen from their cells while receiving nutrition and a home in return.
Today, these early invaders occupy almost all eukaryotic cells, using oxygen to form the primary fuel for life, ATP. During their long occupation at the center of the cellular environment, mitochondria have evolved numerous other functions that are integral to life, for example, regulating cell death and differentiation, cellular responses to stress, as well as calcium balance and immunity.
In fact, we are so reliant on these organelles that problems related to the mitochondria are associated with many disease states including cancer, diabetes, infertility and Alzheimer's disease. Understanding how mitochondria fulfill all of their crucial roles is therefore a key ongoing challenge in biology.
Crucial to their function, the mitochondria contain their own DNA, which is different in structure to the DNA in our nucleus. Representing a remnant of their bacterial past, the mitochondrial genome contains genes important for energy generation.
For a long time, it was thought that the control of the mitochondrial genome was relatively simple, for example with just a few key proteins responsible for the expression of the 37 genes found in human mitochondrial DNA. However, it is increasingly understood that things may not be as simple as they first appear.
Of particular interest to us were two key findings from other research groups: 1) Mitochondrial DNA shows chemical modifications such as methylation, which are known to exist in our main nuclear genome and which regulate the expression of genes. 2) Proteins responsible for the expression of genes in our nuclear genome (transcription factors) can be found in the mitochondria even though they were previously thought to have no function there.
With this in mind, we wanted to investigate which parts of the mitochondrial genome show chemical modifications, how these modifications vary in response to environmental stress and if these modifications are somehow connected to the presence of nuclear transcription factors.
In order to examine chemical modifications in the mitochondrial genome (termed the epigenome), I and my colleagues at Uppsala and Linköping University, Sweden, exposed young chickens to unpredictable lighting conditions during development. This light stress is known to disrupt normal circadian rhythms, causing physiological stress, which we hypothesized would impact the mitochondria.
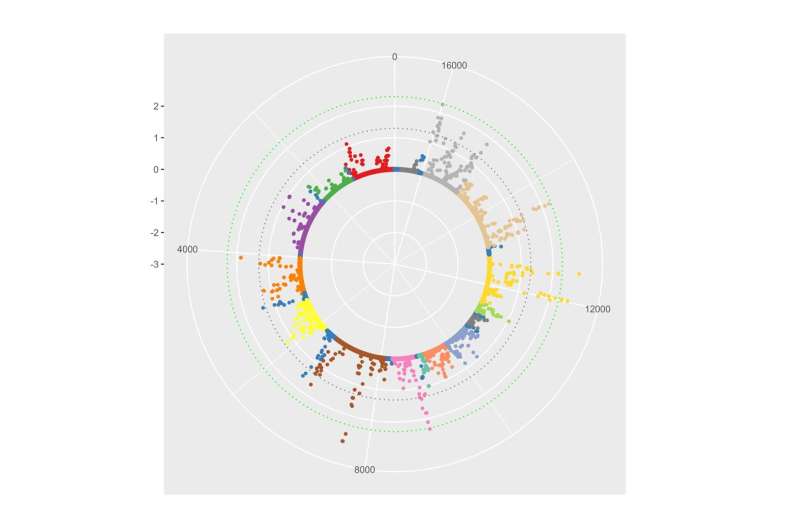
Next, we extracted DNA from the light-responsive center of the birds' brains and used an antibody to bind methylated mitochondrial DNA. By reading the sequence of this methylated DNA in stressed and unstressed male and female birds, we could identify regions of the mitochondrial DNA that were differentially methylated both as a result of circadian disruption and as a result of sex.
As we hypothesized, there were indeed regions of differentially methylated DNA throughout the mitochondrial genome that responded to our stressor. Interestingly, the patterns of methylation were different depending on whether the birds were male or female, indicating that mitochondrial stress responses vary depending on sex.
These results were, however, only one part of the story. Crucial to our study, we wanted to determine whether the regions of methylation might be associated somehow with the nuclear transcription factors that had previously been found lurking within the mitochondria.
By matching the DNA sequences of the methylated regions against a database of sequences associated with binding of transcription factors, we found three hits. Excitingly, two of the matching transcription factors, ATF4 and HNF4A, are central players in known cellular stress responses involving the mitochondria.
Up until now, these transcription factors have been thought to act only on nuclear genes but our data hint at the possibility that they also act directly on the mitochondrial DNA, perhaps in a methylation-dependent manner. If this connection were to be shown experimentally, it would represent a new chapter in our understanding of communication between the nuclear and mitochondrial genomes that drives so many important cellular processes, opening up a new field of mitochondrial biology.
You might ask the question, "But what if this is just the case in chickens?". In order to see whether our findings might be relevant more broadly, we searched for ATF4 and HNF4A binding sites within other diverse mitochondrial genomes, including those of alligators, zebrafish and of course humans. Indeed, regions with similar sequences were found in these species as well, suggesting that the novel mito-nuclear communication that we suggest is present across vertebrates.
Of course, the results that we have found represent just a first step in establishing the nature of methylation-dependent communication between the mitochondrial and nuclear genomes. Our study is now published in BMC Genomics.
With future investigations, we hope to determine the extent of this methylation-dependent mito-nuclear communication with key questions to answer such as "Do nuclear transcription factors bind to the mitochondrial DNA?" "Is this binding affected in some way by the methylation of the DNA?" and "Is the expression of mitochondrial genes affected by any differential binding?"
Right now inside every cell of every person, there is a conversation happening in which the DNA from our parents (in the cell nucleus) is communicating with a small fragment of DNA inside a bacteria which invaded the cells of our ancestors billions of years ago and which is crucial in maintaining cellular function in the face of stress. Often reality is stranger than science fiction.
This story is part of Science X Dialog, where researchers can report findings from their published research articles. Visit this page for information about ScienceX Dialog and how to participate.
More information: John Lees et al, The mitoepigenome responds to stress, suggesting novel mito-nuclear interactions in vertebrates, BMC Genomics (2023). DOI: 10.1186/s12864-023-09668-9
Journal information: BMC Genomics
I am a researcher at Uppsala University, Sweden. My research interests regard metabolism and epigenetics. Specifically, I am using a combination of molecular and physiological techniques to understand the interaction between the mitochondrial and nuclear genomes in response to environmental stressors and how these interactions shape the metabolic phenotype. Through my research I have gained insight from studying unique organisms that are at phenotypic extremes. Although my work focuses primarily on birds, I seek to discover rules that are more broadly applicable.