This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
A new bioimaging method for speeding up and simplifying chemicals identification in tissues
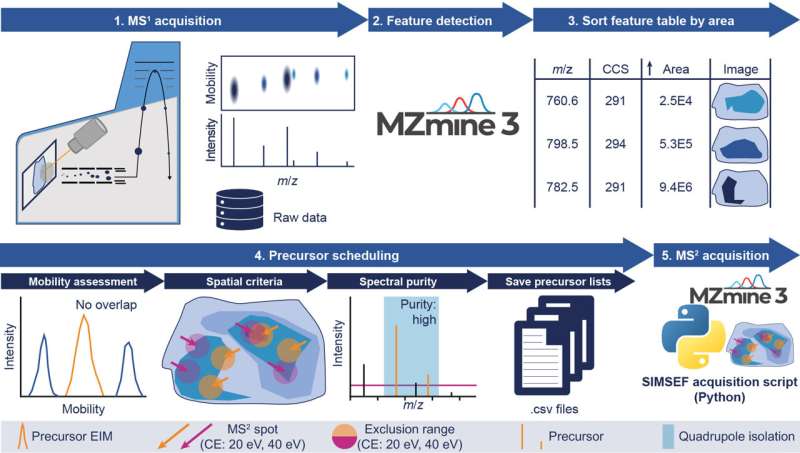
Scientists associated with the international software project MZmine, led by Dr. Robin Schmid and Dr. Tomáš Pluskal from the Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences, have come up with a new piece of software that significantly speeds up and simplifies the identification of chemicals in tissues.
It enables researchers to confidently recognize chemicals and visualize their distribution in organs. Compared to established workflows, the new pipeline requires less work in the laboratory to gain similar insights into the chemistry happening within, for example, tumors and inflammatory lesions. The paper presenting this software to the global scientific community has just been published in the journal Nature Communications.
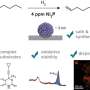
At IOCB Prague, Dr. Robin Schmid is mostly involved in developing the software SIMSEF (spatial ion mobility-scheduled exhaustive fragmentation) together with his colleagues from the group of Prof. Dr. Karst at the University of Münster, Germany, where he was previously based.
The work is done in collaboration with the developers of the timsTOF fleX state-of-the-art mass spectrometer from Bruker Daltonics, which enables the high-level elucidation of the composition of molecules by measuring the mobility of ions.
"Up until now, scientists could tell with a mass spectrometer that this was the formula of the molecule they were looking at, but then when they looked at the database and tried to identify this substance, it was very difficult. The reason is that biological samples can contain numerous distinct lipids and combinations of molecules that are often biologically very different."
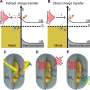
"Now, thanks to the new algorithm, it is possible to look inside the molecule, learn what it is made up of and even compare images with each other," explains Robin Schmid. This can provide important information, for example that a specific part of the brain contains a different type of lipid than another part of this essential organ and that this particular lipid is absent in other tissue.
The SIMSEF algorithm was developed in collaboration with medical experts from German and Swiss universities. Crucially for medical doctors, the detection of clinical biomarkers for diagnostic purposes is also significantly accelerated. Learning very quickly, for example, whether they are dealing with a lipid or metabolite that is only found in cancerous tissue, plays a critical role in deciding on further treatment. Needless to say, this has great implications for its outcome.
The new algorithm is part of the open-source software package MZmine, which has been helping experts worldwide to analyze mass spectrometry data since 2005. Steffen Heuckeroth from the MZmine team and the Institute of Inorganic and Analytical Chemistry at the University of Münster has been the main developer of the SIMSEF algorithm and is the first author of the paper now published in Nature Communications.
He adds, "The article in question does not contain any specific discovery in the field of biology. However, thanks to our new method, many scientists can improve and accelerate their work, which opens up room for new discoveries."
"I always think it's great when we enable other people to improve the working conditions for their research and also to communicate it to other scientists. Thanks to the feedback from the scientific community using MZmine, the program itself directly pushes their research forward," adds Robin Schmid from IOCB Prague.
The director of IOCB Prague Prof. Jan Konvalinka points out, "Everybody is talking about machine learning and artificial intelligence these days. Our colleagues from Dr. Pluskal's team are using it to solve highly specific problems in research as well as in clinical praxis."
The third generation of MZmine, reported in spring 2023 in the journal Nature Biotechnology, can process thousands of samples per hour. The algorithms newly developed by this international team thus further enhance existing capabilities to link different types of data, including combining data from analytical and imaging methods, which until now has not been possible with any academic or commercial software.
More information: Steffen Heuckeroth et al, On-tissue dataset-dependent MALDI-TIMS-MS2 bioimaging, Nature Communications (2023). DOI: 10.1038/s41467-023-43298-9
Journal information: Nature Communications , Nature Biotechnology
Provided by Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences (IOCB Prague)