This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Artificial design and biosynthesis of a single-domain catenated dihydrofolate reductase
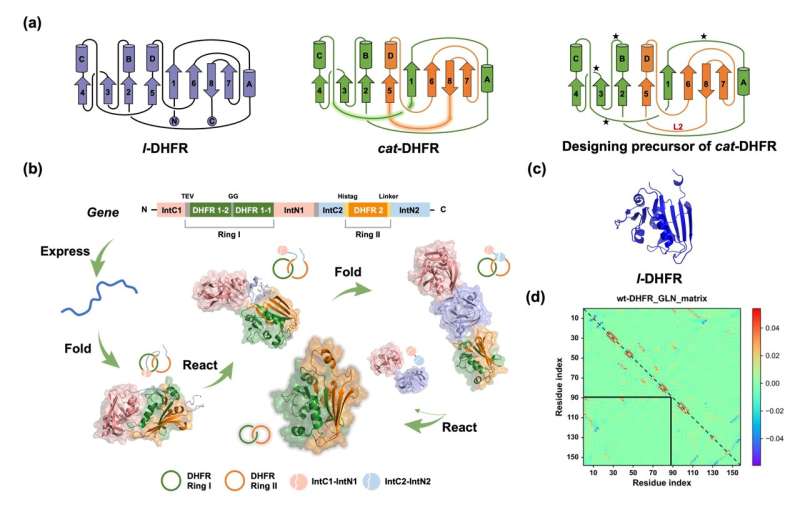
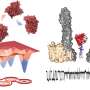
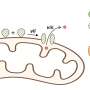
This study was led by Prof. Wen-Bin Zhang (College of Chemistry and Molecular Engineering, Peking University & Beijing Academy of Artificial Intelligence) and Dr. Jing Fang (College of Chemistry and Molecular Engineering, Peking University). A single-domain protein catenane refers to two mechanically interlocked polypeptide rings that fold synergistically into a compact and integrated structure, which is extremely rare in nature.
This design was achieved by rewiring the connectivity between secondary motifs to introduce artificial entanglement, and synthesis was readily accomplished through a series of programmed streamlined post-translational processing events in cells without any additional in vitro reactions.
The single-domain catenane cat-DHFR was thoroughly characterized. Evidence from combined SDS-PAGE, SEC, LC-MS, IMS-MS, and proteolytic digestion experiments unambiguously proved its topology. The cat-DHFR exhibits enhanced anti-aggregation properties and has a Tm that is 6 °C higher than the linear control.
Although the catalytic activity of cat-DHFR is reduced owing to its decreased affinity toward the substrate and cofactor, it has better thermal resilience than l-DHFR. Even after incubation at 70 °C for 10 min, cat-DHFR retained over 70% of the catalytic activity, whereas the linear control lost almost all activity.
The research team anticipates that this method could be generally applicable to other single-domain proteins, including those with folds similar to DHFR or with completely different folds. The availability of these single-domain protein catenanes facilitates the elucidation of topological effects on structure–property relationships.
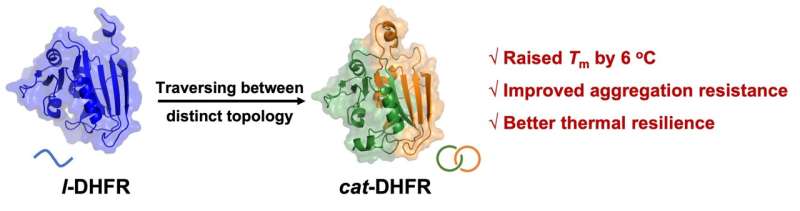
The results further imply that it is possible to map the current linear protein universe into single-domain protein catenanes with well-preserved functions and additional benefits, opening up new territory for protein molecules. Surpassing the linear paradigm of natural protein molecules, these topological proteins are multi-chain, multi-dimensional molecules with functional benefits of topology, rich design possibility, and excellent evolvability.
As a new class of protein molecules, they hold great potential for a broad range of applications, including, but not limited to, industrial enzymes, antibodies, cytokines, and biomaterials.
The study is published in the journal National Science Review.
More information: Jing Fang et al, A Single-domain Protein Catenane of Dihydrofolate Reductase, National Science Review (2023). DOI: 10.1093/nsr/nwad304
Provided by Science China Press