This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Pioneering automated proteoform imaging
Investigators led by Neil Kelleher, Ph.D., professor of Medicine in the Division of Hematology and Oncology and of Biochemistry and Molecular Genetics, have developed an automated technique for imaging and identifying proteoforms in ovarian cancer tissue, according to results published in Nature Communications.
The technique offers the greatest speed and accuracy currently available for the high-resolution, high-throughput imaging of proteoforms—all the modified versions of proteins in a tissue sample—and has multiple potential applications in cancer diagnostics, Kelleher said.
Several techniques are currently used to image proteins in human tissue, but very few are capable of imaging proteoforms. Those that can sample proteoforms directly from tissue do so by ionizing them for mass spectrometry. Other techniques prevent scientists from understanding where the proteoforms exist within a tissue and do not identify all existing proteoforms in a sample at once.
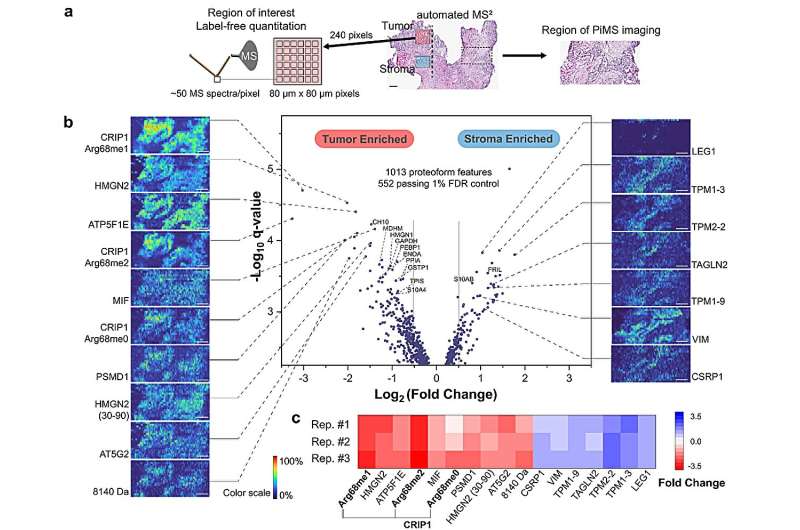
To better understand the spatial position of proteoforms within a given tissue, Kelleher's team developed proteoform imaging mass spectrometry (PiMS), which was detailed in a 2022 study published in Science Advances. The technique works by sampling proteoforms from the tissue with nanodroplets—"weighing" the extracted proteoforms to identify those up to a certain size and then using this data to construct proteoform images of the scanned tissue.
In the current study, Kelleher and his collaborators built upon this technique to create AutoPiMS, which utilizes a computational engine to automatically identify and characterize proteoforms within a thin section of cancer tissue.
AutoPiMS was able to identify and characterize more than 300 proteoforms within a human ovarian cancer tissue sample, and mapped where specific cancer-associated proteins existed within the sample at a speed of one minute per proteoform. AutoPiMS was also able to identify cancer tissues versus non-cancerous tissues from the same patient.
"This technique is really important for cancer tissue imaging and cancer diagnostics," said Kelleher, who also directs the Proteomics Center of Excellence and the Chemistry of Life Processes Institute. "We have shown in this paper that we can locate not just proteins, but their myriad proteoforms, the ultra-specific kind of measurement in my field of proteomics."
The technique will be made available to other Northwestern proteomics investigators, Kelleher said, and he hopes AutoPiMS will accelerate discoveries in the field.
Moving forward, Kelleher and his collaborators will adapt the technique for use in single-cell proteomics, he said.
"If we can have information on single-cell proteoforms, we would have the most precise information about proteins in space, time and composition. That's the ultimate technology that would make protein analysis so much more precise," Kelleher said. "Precision medicine requires precision proteomics. The more precise we can make it, the more we can advance drug development, lower side effects and improve diagnostics, all of which are aligned with the new mission of the CLP Institute at Northwestern."
More information: John P. McGee et al, Automated imaging and identification of proteoforms directly from ovarian cancer tissue, Nature Communications (2023). DOI: 10.1038/s41467-023-42208-3
Journal information: Nature Communications , Science Advances
Provided by Northwestern University