This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
DREAM tool for gene therapies uses 'locally sourced' components
Sourcing some materials closer to home may be a good practice not only in the produce aisle but also the synthetic biology lab.
Seeking to minimize the use of components derived from viral sources, Rice University bioengineers developed a tool that activates silent or insufficiently expressed genes using human-derived proteins known as mechanosensitive transcription factors that naturally enable our cells to turn on specific genes in response to mechanical cues.
Dubbed CRISPR-DREAM (for CRISPR-dCas9 recruited enhanced activation module or DREAM for short), the tool is smaller, more effective and less toxic to medically useful cell types than other state-of-the-art technologies used to control gene expression, according to a study published in Nature Methods.
The DREAM tool could enable better and safer gene and cell therapies and more accurate disease models to address haploinsufficiency disorders, which cause a number of hard-to-treat conditions such as epilepsy and some forms of cancer, immunodeficiency and Alzheimer's disease.
"Many human diseases are driven by problems with too little of a gene being produced," said Isaac Hilton, assistant professor of bioengineering and biosciences at Rice. "You encounter these health issues where people don't make enough of a certain protein or gene product, and in those cases, unfortunately there are often few therapeutic options."
To address this issue, in recent years researchers have used CRISPR-based targeting systems to develop cutting-edge synthetic transcription factors. Most of these tools are built with materials of nonhuman origin, which may come with unwanted side effects.
"One challenge with some of the current technologies is that they are built using viral proteins that have evolved to reprogram how our cells work, and they do so in ways that are of course not necessarily beneficial," Hilton said. "And even though virally derived elements can be engineered to work for the host cell's benefit, we and others have observed in our research that they can still cause some toxicity in human cells."
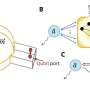
Instead of relying on virally derived proteins, Barun Mahata, a postdoctoral researcher in Hilton's lab, sought to employ transcription factors that human cells already make and use. Mahata fused the specific parts of these proteins responsible for activating genes to CRISPR-based programmable delivery platforms using a method intended to enhance their transcriptional powers.
"We harnessed the natural ability of human-derived transcription factors, or proteins responsible for gene synthesis in the cells of our body," said Mahata, who is the lead author on the study. "The transcription activation units we built function in a very precise way. They induce gene activity where we target them and can activate very rapid and robust transcription."
They also help tackle another disadvantage of current synthetic gene-activation platforms, which are often too large for efficient delivery to human cells.
"What we did was look for small segments of human proteins that we could leverage to apply these technologies in human cells more effectively," said Hilton, the corresponding author on the study and a Cancer Prevention and Research Institute of Texas (CPRIT) scholar. "When we started this project of building synthetic transcription factors with human-derived proteins, we wanted to identify the ideal source material with compactness being one key factor."
Human mechanosensitive transcription factors—proteins that our cells and organs use to respond to mechanical forces—met the researchers' criteria: In addition to being relatively small, they are quick-acting and widely used by nearly all human cell types.
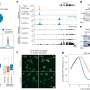
Moreover, the team was able to activate up to 16 different locations on the genome simultaneously ⎯ a record number for synthetic transcription factors.
"The reason why that capability is particularly important is because when our cells perform a function, it's not just that they turn on a single gene," Hilton said. "Instead, they typically turn on whole constellations or networks of genes in concert. And we can now use these synthetic transcription factors to mimic and engineer what our cells do naturally."
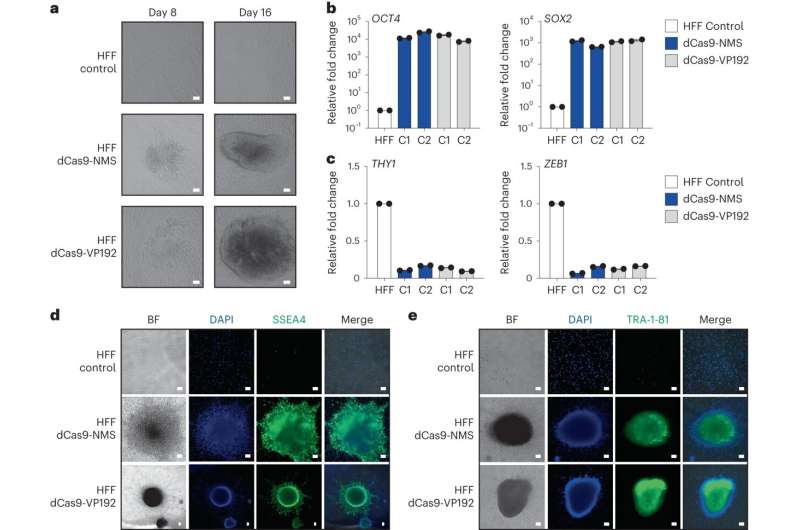
The researchers showed that the DREAM tool could be used to convert skin cells to induced pluripotent stem cells (iPSCs) in a dish as a proof of concept exercise—a feat that Hilton says carries "tremendous biomedical utility" in the long run.
More information: Mahata, B. et al. Compact engineered human mechanosensitive transactivation modules enable potent and versatile synthetic transcriptional control, Nature Methods (2023). DOI: 10.1038/s41592-023-02036-1 www.nature.com/articles/s41592-023-02036-1
Provided by Rice University