This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Scientists discover a new enzyme that helps cells fight genomic parasites
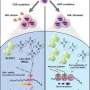
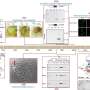
![Identification of the catalytic center of TOFU-2. a, Model of piRNA (21U RNA) formation in C. elegans. Individually transcribed piRNA precursors are stabilized by PETISCO. After the removal of the 5′-cap and two nucleotides, intermediates are loaded onto PRG-1, followed by trimming and 3′-end methylation. The nuclease that processes the 5′-end is currently unclear. b, Schematic of TOFU-1, TOFU-2 and SLFL-3/4, in comparison to rat SLFN13. The lines indicate low-complexity regions and the rectangles indicate the predicted folded domains. BD, bridging domain. c, Superposition of TOFU-1 and TOFU-2 SLFN domains onto the crystal structure of the N-terminal SLFN13 endoribonuclease domain (Protein Data Bank (PDB): 5YD0). Domains are colored as in b. The magnified view shows the active site of SLFN13. Involved residues are shown as sticks. d, Label-free proteomic quantification of TOFU-2–HA and wild-type immunoprecipitates from young adult extracts. n = 4 biological replicates. The x axis shows the median fold enrichment of individual proteins, and the y axis shows −log10[P]. P values were calculated using Welch two-sided t-tests. The dashed lines represent enrichment thresholds at P = 0.05 and fold change > 2, curvature of enrichment threshold c = 0.05. The dots represent enriched (blue/red) or quantified (gray) proteins. Only uniquely matching peptides were used. e, Schematic of the mCherry–H2B piRNA sensor. f, Wide-field fluorescence microscopy analysis of adult hermaphrodites carrying the piRNA sensor in the following three genetic backgrounds: tofu-2(E216A) (top), prg-1(n4357) (middle) and wild type (bottom). Germlines are outlined by white dashed lines. Scale bar, 50 µm. A representative image from a series of ten is shown. g, Total mature piRNA levels (type 1) in wild-type and tofu-2(E216A)-mutant young adult hermaphrodites. n = 3 biological replicates. The red lines show the group means. P values were calculated using two-tailed unpaired t-tests. h, The relative abundance of type 1 piRNA precursors from individual loci in tofu-2(E216A)-mutant versus wild-type young adult hermaphrodites. n = 3 biological replicates. RPM, reads per million non-structural small RNA reads. Credit: Nature (2023). DOI: 10.1038/s41586-023-06588-2 Scientists discover a new enzyme that helps cells fight genomic parasites](https://scx1.b-cdn.net/csz/news/800a/2023/scientists-discover-a-6.jpg)
The research teams of Professor René Ketting at the Institute of Molecular Biology (IMB) in Mainz, Germany, and Dr. Sebastian Falk at the Max Perutz Labs in Vienna, Austria, have identified a new enzyme called PUCH, which plays a key role in preventing the spread of parasitic DNA in our genomes. These findings may reveal new insights into how our bodies detect and fight bacteria and viruses to prevent infections.
Our cells are under constant attack from millions of foreign intruders, such as viruses and bacteria. To keep us from getting sick, our bodies have an immune system—a whole army of cells that specializes in detecting and destroying these invaders. However, our cells face threats not only from external enemies but also from within.
Genomic parasites populate a large part of the genome
An amazing 45 percent of our genome is comprised of thousands of genomic parasites, i.e., repetitive DNA sequences called transposable elements (TEs). TEs are found in all organisms but have no specific function. They can, however, be dangerous. TEs are also called "jumping genes" because they can copy and paste themselves into new locations in our DNA.
This is a major problem because it can lead to mutations that cause our cells to stop working normally or to become cancerous. As such, almost half of our genome is engaged in a constant guerrilla war with the other half as TEs seek to multiply, while our cells try to prevent them from spreading.
How do our cells combat these internal enemies? Fortunately, our cells have evolved a genomic defense system of specialized proteins whose job it is to hunt down TEs and prevent them from replicating.
In a new paper published in Nature, René Ketting and Sebastian Falk together with their research teams report their discovery of PUCH—a completely new, previously unknown type of enzyme, which is key to this genomic defense system. They found that PUCH plays a crucial role in producing small molecules called piRNAs, which detect TEs when they attempt to "jump." They then activate the genomic defense system to stop TEs before they paste themselves into new locations in our DNA.
The researchers discovered PUCH in the cells of the roundworm C. elegans, a simple invertebrate often used in biological research. However, the findings may also shed light on how our own immune system works. PUCH is characterized by unique molecular structures called Schlafen folds.
Enzymes with Schlafen folds are also found in mice and humans, where they appear to play a role in innate immunity, the body's first line of defense against viruses and bacteria. For example, some Schlafen proteins interfere with the replication of viruses in humans. On the other hand, some viruses such as monkeypox viruses, for example, may also use Schlafen proteins to attack the cell's defense system. René Ketting suspects that Schlafen proteins may have a wider, conserved role in immunity in many species, including humans.
"Schlafen proteins may represent a previously unknown molecular link between immune responses in mammals and deeply conserved RNA-based mechanisms that control TEs," said Ketting, who is also a Professor of Biology at Johannes Gutenberg University Mainz (JGU). If so, Schlafen proteins may represent a common defense mechanism against both external enemies like viruses and bacteria as well as internal ones such as TEs.
"It's conceivable that Schlafen proteins have been repurposed into enzymes that protect cells from infectious DNA sequences, such as TEs," added Sebastian Falk. "This discovery may profoundly impact our understanding of innate immune biology."
More information: Nadezda Podvalnaya et al, piRNA processing by a trimeric Schlafen-domain nuclease, Nature (2023). DOI: 10.1038/s41586-023-06588-2
Provided by Universitaet Mainz