This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Modulation of protein stability: A new approach to studying cosolvent effects
Controlling the process of destabilization is important when manipulating the unfolding and refolding of proteins in vitro (outside their native environment). To this end, urea and alcohol are used as cosolvents, substances added in small amounts along with water, to destabilize and denature proteins.
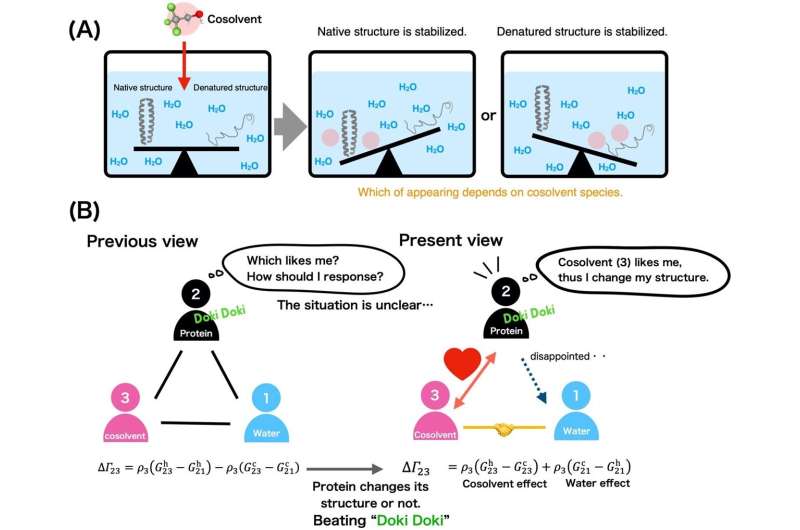
Urea disturbs a native protein to produce disordered coils, and the interference by alcohol treatment yields helical structures. Research on the mechanism of cosolvents has shown that a protein's stability between its native and denatured states is tied to how the cosolvent binds to each state.
Binding of a cosolvent to a protein's native or denatured state is governed by preferential binding parameters (PBPs)–specific interactions with the protein. Unfortunately, the molecular mechanisms affecting protein stability in the presence of a cosolvent are not fully understood.
But is it possible to pinpoint the differences between how cosolvents like urea and alcohol interact with a protein? A team of researchers from Japan has recently addressed this question using a tool that explains how cosolvents interact with a protein and predicts the all-important binding relationship between the protein, water, and cosolvent.
"We wanted to gain a deeper insight into how a cosolvent stabilizes a protein's native or denatured state in solution. So, we leveraged computational simulations to determine the PBPs and reveal how urea and 2,2,2-trifluoroethanol (TFE) induced their opposing cosolvent effects," says team leader Tomonari Sumi, Associate Professor at the Research Institute for Interdisciplinary Science at Okayama University.
Additionally, Ms. Noa Nakata and Dr. Kenichiro Koga from Okayama University, Dr. Ryuichi Okamoto from the University of Hyogo, Dr. Takeshi Morita from Chiba University, and Dr. Hiroshi Imamura from Nagahama Institute of Bio-Science and Technology were involved in the research and served as co-authors in this study published in Protein Science.
The team studied the cosolvent-dependent stability between native and denatured states in the yeast protein, GCN4-p1, which has a well-defined native structure that consists of coils and helices. This made it ideal for studying TFE's helix stabilization and urea's coil stabilization in the presence of water using molecular dynamics simulations, a computational method to predict the movement of atoms in a protein over a given time.
"We were able to predict the suitable excess preferential binding (EPB) for both cosolvents. First, electrostatic interactions between TFE and the side chains of GCN4-p1's helices yielded the EPB, stabilizing the helices. Second, dispersion and electrostatic interactions between urea and the coils of GCN4-p1's main chains contributed to the EPB, stabilizing the coils" explains Dr. Sumi.
Furthermore, the group's simulation data corroborated with experimental data on helix and coil stabilization for TFE and urea. In this way, the team demonstrated that these interactions produced opposing cosolvent effects. In the context of the triangular protein-cosolvent-water relationship, hydroxyl groups of TFE were drawn to the protein's polar side chains, while urea preferentially bound to the protein's peptide backbone. This was a significant finding as one can effectively predict the potential structural changes that a cosolvent may induce.
The team is excited by the broader implications of their research as it solves the problem of cosolvent screening by leveraging computational resources. "We're confident these findings will bear fruit for protein stabilization across industry and medicine. The analytical methods applied in this study significantly reduce the time needed to understand how cosolvents influence a protein's structural stability. This can help us bypass the more traditional methods that involve time-consuming free energy calculations," concludes Dr. Sumi.
More information: Noa Nakata et al, Molecular mechanism of the common and opposing cosolvent effects of fluorinated alcohol and urea on a coiled coil protein, Protein Science (2023). DOI: 10.1002/pro.4763
Provided by Okayama University