This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
When predicting the function of microbial communities, simpler may be better
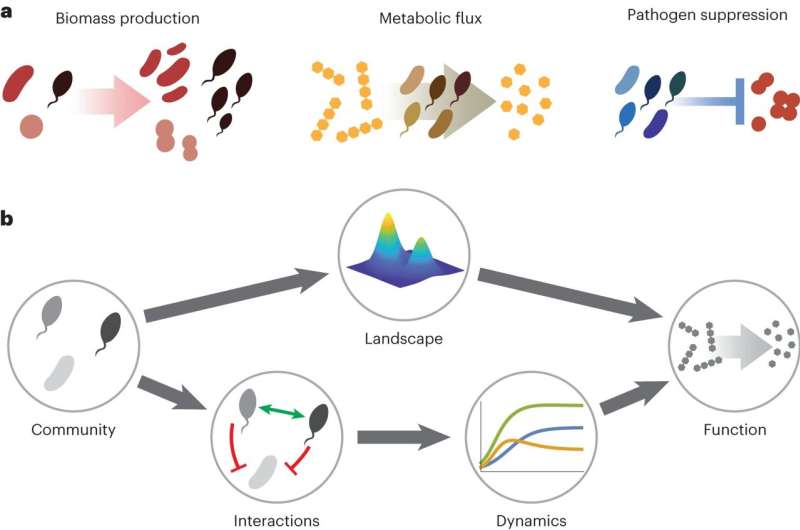
As biologists learn more about how microbial communities work together, a major goal is understanding how their composition determines function. What combination of strains and species makes the best team for breaking down pollutants, for example, or fighting off infections? For years, scientists attempted to crack this problem with calculations about how microbial species interact with each other, but the nearly endless potential combinations of microbes in each community render these calculations nearly impossible.
A new study by researchers from the University of Chicago, Yale University, and Washington University in St. Louis shows that taking a simpler approach may be better. In a paper published in Nature Ecology & Evolution on October 2, 2023, they demonstrate a statistical model that can accurately predict functions of a microbial community just by considering the presence or absence of different species and not the details of their interactions.
This approach worked across a variety of datasets from different ecological systems, suggesting that it could be useful for designing microbial communities with a specific function for many different applications.
"Predictions using the standard approaches are themselves very challenging and require a lot of data," said Seppe Kuehn, Ph.D., Assistant Professor of Ecology and Evolution at UChicago and co-senior author of the study. "In some cases that approach has been successful. But what's quite surprising in our paper is that we're just as successful if we ignore all of that."
The researchers used a concept borrowed from genetics. The way in which genetic mutations impact an organism's fitness, or ability to survive and pass genetic material to offspring, is commonly depicted as a "fitness landscape." In this picture, different combinations of mutations are conceptualized as points on an abstract "map" of possible organisms; the height of the landscape above each point corresponds to the fitness of that organism. The result looks like a topographical map with peaks and valleys representing high- and low-fitness organisms.
In the new study, instead of fitness landscapes, the researchers considered an analogous landscape of community function, where adding or removing species is akin to "mutations," and the topographical peaks represent communities with, say, a high production rate of some compound.
In principle, the shape of such landscapes could be arbitrarily complex, or "rugged," with many peaks and valleys. But testing this approach on six different datasets from different labs, the team discovered that the landscapes were surprisingly smooth. This smoothness meant the shape of the landscape could be approximated with relatively little data, allowing the researchers to predict community function just as well as the more complicated approaches that account for species dynamics and abundance.
The simplicity of the approach may contribute to its robustness. "The model worked well compared to other statistical approaches, but more importantly, it seems to work consistently well across different datasets that have very different microbes for different functions," said Abby Skwara, the lead author of the study, who was an undergraduate at UChicago and is now a graduate student at Yale University.
Among the six examples tested by the researchers, one measured the ability of microbial communities to produce butyrate, a short-chain fatty acid that is important for healthy digestion. Another dataset measured the breakdown of starch.
The researchers hope that this new landscape model can provide a tool to help design microbial communities for a specific purpose, like breaking down environmental contaminants in soil, or producing the right metabolites to help restore healthy digestive systems. In the process, it could also lead to a greater understanding of how microbial communities function in the first place.
"The success of this simple approach is intriguing in that it contradicts our intuition about ecological complexity," said Mikhail Tikhonov, Ph.D., Assistant Professor of Physics at WashU and co-senior author. "Here, the communities are complex, yet the landscapes of their function are not. Understanding why this is the case is an exciting question for theory."
More information: Abigail Skwara et al, Statistically learning the functional landscape of microbial communities, Nature Ecology & Evolution (2023). DOI: 10.1038/s41559-023-02197-4
Journal information: Nature Ecology & Evolution
Provided by University of Chicago