This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Examining the genesis of CRISPR's molecular scissors
Genome engineering may be the future of medicine, but it relies on evolutionary advances made billions of years ago in primordial bacteria, the original masters of gene editing.
Modern day genome engineers like Columbia's Sam Sternberg are always looking forward, modifying these ancient systems and pushing them to perform ever more complex feats of gene editing.
But to uncover new tools, it sometimes pays to look backward in time to understand how bacteria first created the original systems, and why.
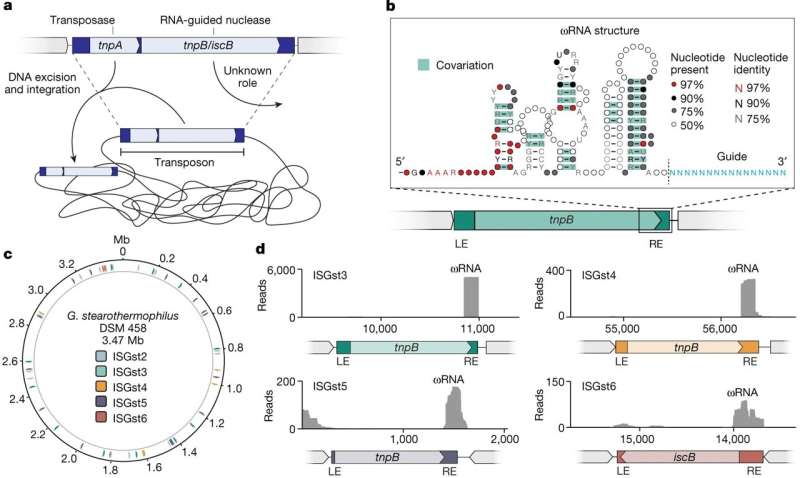
In a new study, published in Nature, Sternberg and his postdoc, Chance Meers, Ph.D., took a trip back in time to look at the precursors of CRISPR-Cas9—which lurk inside so-called "jumping genes"—to unravel how CRISPR's DNA scissors evolved.
Their findings reveal how thousands of newly discovered DNA scissors work and how they might be engineered into new genome engineering technologies.
CRISPR-Cas9 comes from jumping genes
Within bacteria, CRISPR-Cas9 plays a crucial role in protecting cells from viral infections. Using the help of a guide RNA, these molecular scissors first recognize the DNA of invading viruses and then slice apart the viral genome.
A few years ago, scientists traced CRISPR-Cas9's origins to transposons, mobile genetic elements, also known as "jumping genes," that hop from one location in the genome to another through an enigmatic process known as transposition.
"A lot of the biological topics we study in our lab arose when one life form stole genes from another—for example, a bacterium stealing genes from a mobile genetic element, like a virus, plasmid, or transposon—and then those genes were repurposed to perform similar biochemical reactions, but for a different function altogether," Sternberg says.
The transposon link soon led researchers to a treasure trove of potential new editing tools: thousands of ancient transposons that are still active in bacterial genomes, each carrying an RNA-guided DNA nuclease that could potentially be programmed by genomic engineers—the human kind—to cut DNA.
Genome engineers are now working to exploit these systems, but to Sternberg and Meers, one important question was still unanswered.
"These transposons jump into and out of the genome with the help of their own enyzmes, called transposases," Meers says. "They don't need DNA scissors, and they don't need guide RNAs either.
"So why do they carry the genes for RNA-guided DNA scissors?"
Without DNA scissors, transposons would go extinct
It was a tough problem to crack.
The first hurdle Meers solved was finding the right bacteria with many copies of active transposons, to serve as a model system. The common lab bacterium, E. coli, wasn't the best starting point, so Meers chose Geobacillus stearothermophilus, a heat-loving bacteria with dozens of active jumping genes.
Meers also studied the problem from the perspective of the transposon, developing powerful assays that captured the jumping genes in the process of moving around the bacterial genome, hopping in and out of plasmids, and from one bacterial strain to another. "Without that approach, you end up just studying the DNA scissors in isolation, preventing a holistic view that pieces the whole story together," Sternberg says.
With these assays in hand, Meers and Sternberg—aided by a team of colleagues within the lab—dove deep into the way transposons moved around and showed that without the DNA-cutting scissors, the jumping genes could hop into new locations but were prone to very rapid extinction. (Bacteria continually try to inactivate mobile genetic elements, including transposons.)
The CRISPR-like molecular scissors prevented extinction by guiding a copy of the transposon back to the location it jumped from, after cutting the DNA to slip the copy into place.
"With this 'cut and copy' strategy, the transposon can proliferate at a rate faster than its rate of permanent loss," Meers says. In effect, nature's most powerful genome editors originally evolved to edit themselves into the genome, selfishly promoting their own spread.
More versions of CRISPR to find?
Because CRISPR-Cas evolved from a transposon found in hundreds of thousands of copies across the bacterial kingdom, it's likely that nature has created other systems from these powerful transposon genes that are waiting to be discovered.
"It's hard to believe that evolution stopped inventing molecular scissors with the genetics of CRISPR-Cas," Sternberg says. "There must be other systems at work, and if we find them, we may be able to borrow those genes as well and use them for another purpose: engineering the genome of human cells."
More information: Samuel Sternberg, Transposon-encoded nucleases use guide RNAs to bias their inheritance, Nature (2023). DOI: 10.1038/s41586-023-06597-1. www.nature.com/articles/s41586-023-06597-1
Journal information: Nature
Provided by Columbia University Irving Medical Center