This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
New algorithm for quicker detection of antibiotic resistances
A team of researchers led by microbiologist Professor Dr. Axel Hamprecht of the University Medicine Oldenburg has developed a new method that enables quicker detection of a frequently overlooked antibiotic resistance. The team focused on determining whether a strain of the bacterium Proteus mirabilis is resistant to certain antibiotics, thus rendering them ineffective. Infection with this bacterium can lead to urinary tract infections, wound infections, bloodstream infections and pneumonia. The researchers have now presented their innovative technique in the journal Clinical Microbiology and Infection.
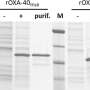
The team was able to demonstrate that resistances to reserve antibiotics (last-resort antibiotics) in Proteus mirabilis are far more common than previously believed, and often go undetected with current standard testing methods. To identify these resistances, the group used cutting-edge molecular biology methods including next-generation sequencing, with which many hundreds of genes can be sequenced in parallel.
On the basis of these data the researchers were able to develop a method for the fast and cost-effective detection of resistance to a class of reserve antibiotics called carbapenems. These resistances generally coincide with resistance to a number of other drugs. The Oldenburg researchers' method involves first testing the susceptibility of a bacterial strain to two specific antibiotics that the study showed to be particularly reliable indicators.
In many cases the result of these tests, in conjunction with an algorithm especially developed by the team, can already determine whether the bacterial strain under examination is capable of destroying a carbapenem antibiotic. Armed with this knowledge, doctors can prescribe a different medication if necessary. If the results of the test are unclear, an additional test will be necessary, which the team also described in the paper.
"This approach is certainly considerably faster and cheaper than the current methods for detecting these resistances," says Professor Hamprecht, director of the University Institute of Medical Microbiology and Virology at the Klinikum Oldenburg and a lecturer at the University's School of Medicine and Health Sciences. Another advantage, he adds, is that most laboratories already have the infrastructure for conducting these tests.
Resistances in Proteus mirabilis strains have often gone undetected in the past as they are frequently caused by enzymes that are not tested for in standard testing procedures or most molecular biology methods.
More information: Axel Hamprecht et al, Proteus mirabilis—analysis of a concealed source of carbapenemases and development of a diagnostic algorithm for detection, Clinical Microbiology and Infection (2023). DOI: 10.1016/j.cmi.2023.05.032
Provided by University of Oldenburg