This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Genomic analysis reveals dead-end hybridization between kiwifruit species
Compared to allopolyploid speciation, there are fewer cases of homoploid hybrid speciation in plants. Although transient homoploid hybridization events have been detected in many plant genera, solid evidence from genomic data is scare.
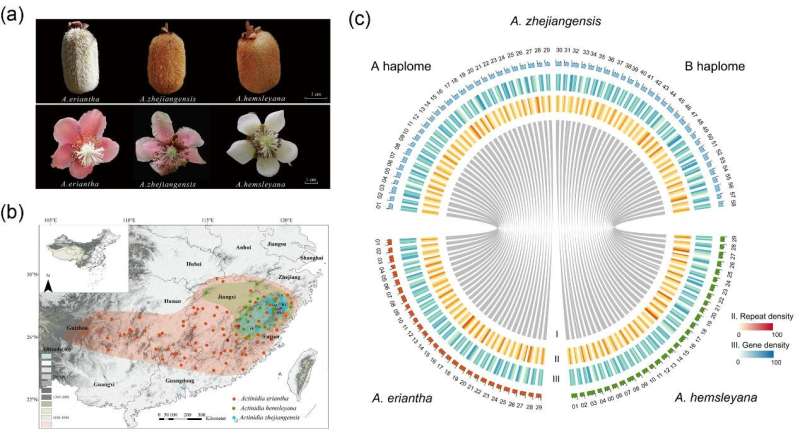
The reticulate evolution caused by interspecific hybridization has greatly contributed to speciation of Actinidia. Actinidia zhejiangensis, a species published in 1982, is distributed in Jiangxi, Zhejiang and Fujian Province in China with a small population size.
Previous studies speculated that A. zhejiangensis originated from hybridization between Actinidia eriantha and Actinidia hemsleyana or Actinidia rufa based on limited molecular markers and small number of sampled taxa. The hybridization origin of A. zhejiangensis, and the phylogenetic relationship between it and other Actinidia species still cannot be determined from the morphological characteristics and the reported molecular evidence.
Researchers from the Wuhan Botanical Garden of the Chinese Academy of Sciences, together with collaborators from Lushan Botanical Garden and Forest Resources Monitoring Center of Zhejiang Province, generated chromosome-scale reference genome assemblies of A. zhejiangensis and A. hemsleyana to reveal the transient homoploid hybridization of A. zhejiangensis.
Results were published in The Plant Journal, titled "Genomic analyses reveal dead-end hybridization between two deeply divergent kiwifruit species rather than homoploid hybrid speciation."
The chromosomes of A. zhejiangensis were confidently assigned to two sets of haplomes. Combined with a published A. eriantha genome, the researchers found these two haplomes originated from A. eriantha and A. hemsleyana, respectively. Based on resequencing data from A. zhejiangensis, A. eriantha, and A. hemsleyana individuals, they discovered that A. zhejiangensis were mainly F1 hybrids of A. hemsleyana and A. eriantha, and A. hemsleyana and A. eriantha were the constant paternal and maternal parents, respectively.
Thus, A. zhejiangensis was not a stabilized independent hybrid species, although gene flow started about 0.98 million years ago, suggesting strong reproductive barriers between A. hemsleyana and A. eriantha.
Five inversions containing genes involved in pollen germination and pollen tube growth could affect the fertility of hybrids between A. hemsleyana and A. eriantha. Despite its distinct morphological traits and long recurrent hybrid origination, A. zhejiangensis did not initiate speciation.
This study provides the first evidence of dead-end hybridization between two deeply divergent kiwifruit species rather than homoploid hybrid speciation, sheds light on Acitinida speciation, and generates valuable genomic resources for future evolutionary and functional genomics studies of kiwifruit.
More information: Xiaofen Yu et al, Genomic analyses reveal dead‐end hybridization between two deeply divergent kiwifruit species rather than homoploid hybrid speciation, The Plant Journal (2023). DOI: 10.1111/tpj.16336
Journal information: The Plant Journal
Provided by Chinese Academy of Sciences




















