This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Genome editing used to create disease-resistant rice

Researchers from the University of California, Davis, and an international team of scientists have used the genome-editing tool CRISPR-Cas to create disease-resistant rice plants, according to a new study published in the journal Nature June 14.
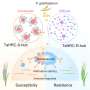
Small-scale field trials in China showed that the newly-created rice variety, developed through genome editing of a recently discovered gene, exhibited both high yields and resistance to the fungus that causes a serious disease called rice blast. Rice is an essential crop that feeds half of the world's population.
Guotian Li, a co-lead author of the study, initially discovered a mutant known as a lesion mimic mutant while working as a postdoctoral scholar in Pamela Ronald's lab at UC Davis. Ronald is co-lead author and Distinguished Professor in the Department of Plant Pathology and the Genome Center.
"It's quite a step forward that his team was able to improve this gene, making it potentially useful for farmers. That makes it important," Ronald said.
The roots of the discovery began in Ronald's lab, where they created and sequenced 3,200 distinct rice strains, each possessing diverse mutations. Among these strains, Guotian identified one with dark patches on its leaves.
"He found that the strain was also resistant to bacterial infection, but it was extremely small and low yielding," Ronald said. "These types of 'lesion mimic' mutants have been found before but only in a few cases have they been useful to farmers because of the low yield."
Working with CRISPR
Guotian continued the research when he joined Huazhong Agricultural University in Wuhan, China.
He used CRISPR-Cas9 to isolate the gene related to the mutation and used genome editing to recreate that resistance trait, eventually identifying a line that had good yield and was resistant to three different pathogens, including the fungus that causes rice blast.
In small-scale field trials planted in disease-heavy plots, the new rice plants produced five times more yield than the control rice, which was damaged by the fungus, Ronald said.
"Blast is the most serious disease of plants in the world because it affects virtually all growing regions of rice and also because rice is a huge crop," Ronald said.
Future applications
The researchers hope to recreate this mutation in commonly grown rice varieties. Currently they have only optimized this gene in a model variety called Kitaake that is not grown widely. They also hope to target the same gene in wheat to create disease-resistant wheat.
"A lot of these lesion mimic mutants have been discovered and sort of put aside because they have low yield. We're hoping that people can go look at some of these and see if they can edit them to get a nice balance between resistance and high yield," Ronald said.
More information: Gan Sha et al, Genome editing of a rice CDP-DAG synthase confers multipathogen resistance, Nature (2023). DOI: 10.1038/s41586-023-06205-2
Journal information: Nature
Provided by UC Davis